Synthetic biology of long-range gene regulation
Prof. Dr. Stefan Mundlos
The genome is organized in a specific three-dimensional (3D) configuration which has a major influence on gene regulation. HiC has shown that mammalian genomes are organized in distinctly folded chromatin modules, called topologically associated domains (TADs) that are separated from each other by boundary regions. TADs can be seen as regulatory units/landscapes that restrict the contacts enhancers can establish with their target genes thus defining regulatory landscapes. We have shown the importance of this concept in disease (Lupianez et al. 2015), but can also be the origin of evolutionary novelty (Real et al. 2020). In an evolutionary context changes in TAD-landscapes need to be incorporated in the existing regulatory context (Ringel et al. 2022). These findings provide a framework for interpreting the effect of structural variations in a disease and evolutionary context.
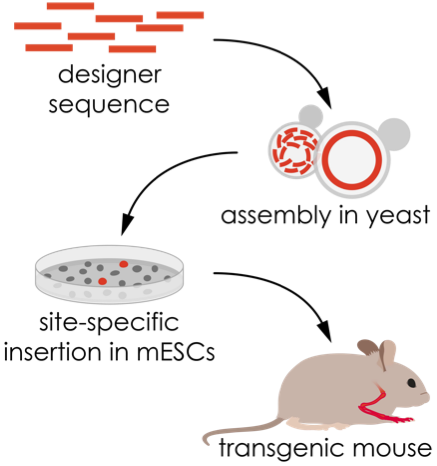
Based on our previous findings we now want to combine cutting-edge genomic engineering with synthetic genomics to manipulate the mouse genome at an unprecedented scale. TADs and regulatory landscapes in general cover large regions of the genome. In vitro cellar systems and/or short artificial constructs are inherently incapable of modelling the full functionality of such landscapes in developmental gene regulation in its full complexity. Genomic engineering and the de novo synthesis of large (up to 150 kb) DNA fragments using yeast assembly approaches make it possible to re-engineer complex rearrangements and entire regulatory landscapes. Technologies for single copy targeting and CRISPR-Cas9 genome editing allow the insertion of these synthetic DNA fragments into mouse embryonic stem cells (mESCs) to produce mice that carry altered regulatory sequences in their genome. This approach's modularity and versatility will help identify and characterize the regulatory effect of individual components and how they work together, giving fundamental insights into gene regulation and how differences in sequence give rise to morphological adaptation and diversity.
References
For more information have a look at the website of the Development & Disease group.
![[NEW] - lncRNAs in 3D – dissecting the gene regulatory function of long non-coding RNAs](/4655620/teaser-1697451260.jpg?t=eyJ3aWR0aCI6MzYwLCJoZWlnaHQiOjI0MCwiZml0IjoiY3JvcCIsImZpbGVfZXh0ZW5zaW9uIjoianBnIiwib2JqX2lkIjo0NjU1NjIwfQ%3D%3D--36effda3255a9e519ace3a72333e319bc6dea078)










