Mechanisms of transcription and RNA processing regulation
Mayer Lab
Introduction & Scope of our research
Gene transcription and RNA processing are the two key steps that can lead to a functional RNA and represent a major target for the regulation of cell function. They also underlie cell growth and differentiation. Dysregulation of transcription and RNA processing is a hallmark of human disease.
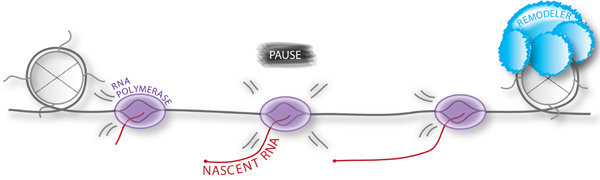
The key enzyme that pervasively transcribes eukaryotic genomes and gives rise to all protein-coding and many non-coding RNAs in the nucleus is RNA polymerase II (Pol II). For decades, Pol II transcription was thought to be regulated primarily during initiation, when RNA polymerase is recruited to the promoter and begins to synthesize a nascent RNA. However, recent advances in sequencing techniques that provide a quantitative measure of nascent RNA and reveal the genomic localization of RNA polymerases at nucleotide resolution have challenged this long-held view. These high-resolution genomic approaches have not only uncovered novel transcriptional activities, but have also provided evidence for widespread transcriptional pausing of Pol II during transcription elongation and termination, suggesting that these later stages of nascent transcription represent important hubs for gene regulation. The molecular mechanisms underlying these post-initiation regulatory events and how transcription is coordinated with RNA processing, including splicing, alternative splicing, and 3'-end RNA processing, to control gene expression are poorly defined. This lack of knowledge is particularly acute in the dynamic context of cell differentiation and disease states.
The goal of our research
The primary goal of our research is to elucidade the key mechanisms that control gene regulation, in particular nascent transcription and RNA processing, in the native chromatin environment of mature and differentiating mammalian cells. Specifically, we seek to understand how post-initiation regulatory events are established in a dynamic chromatin environment in cells and how these regulatory mechanisms control cell differentiation. We are also interested in how a dysregulation of nascent transcription and RNA processing causes human diseases such as cancer.
Our toolbox
We address these fundamental questions through an interdisciplinary combination of high-resolution genome-wide approaches, genome engineering techniques, biochemical methods, bioinformatics tools, and super-resolution microscopy. We are also developing new quantitative methods, including transcriptome-wide high-resolution nascent RNA analysis approaches, to study the molecular mechanisms that control nascent RNA synthesis and RNA processing in mammalian cells.




