
Genome Regulation
Meissner Lab
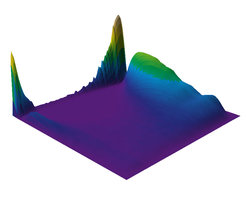
This plot shows the dynamic methylation landscape of the human genome, where the x axis (left) corresponds to the maximal observed methylation change across 24 human cell and tissue types, y is the median total methylation and z is the density of CpG dinucleotides. The methylation of cytosine, usually at CpGs, is a common feature of epigenetic regulation of gene expression. Most cell types have relatively stable CpG dinucleotide methylation patterns and our understanding of which CpGs participate in genomic regulation is still relatively limited.
The Genome Regulation Group is a mixed group of experimental and computational biologists that uses genomic tools to study developmental and stem cell biology with a particular interest in the role of epigenetic regulation. Their work utilizes early mouse development as well as various mouse and human stem cell paradigms to dissect basic molecular mechanisms including the role of DNA methylation and other epigenetic modifications in gene and genome regulation.












