Molecular mechanisms of genome transcription regulation & dysregulation
Prof. Dr. Helge Ewers & Dr. Andreas Mayer
If you want to work at the exciting interface of high-resolution functional genomics and super-resolution microscopy to uncover key regulatory principles that underlie genome transcription in the natural chromatin environment of normal human cells and upon disease states, send your application materials to the IMPRS-BAC and join the Research Groups of Helge Ewers and Andreas Mayer.
The research group of Helge Ewers develops microscopy assays and image analysis routines for the investigation of cellular processes at the mesoscale using super-resolution and single-molecule approaches. To this end, they optimize sample preparation, labeling and selection of microscopy approaches to suit the cell biological problem at hand. Originally interested in the plasma membrane-cytoskeleton interaction, they recently ventured into a new frontier, the organization of nuclear domains.
The Max Planck Research Group “High-Resolution Functional Genomics” of Andreas Mayer studies the regulatory principles that drive genome transcription and RNA processing in human cells, representing the two major steps that can lead to functional RNAs. The group has also a strong interest in how alterations of transcription and RNA processing cause human diseases. To reveal the molecular mechanisms of transcription and RNA processing control in cells, the Mayer lab develops and applies new quantitative genome-wide approaches in combination with computational analysis tools.
Sample project: Determine the regulatory crosstalk between enhancers and target gene transcription
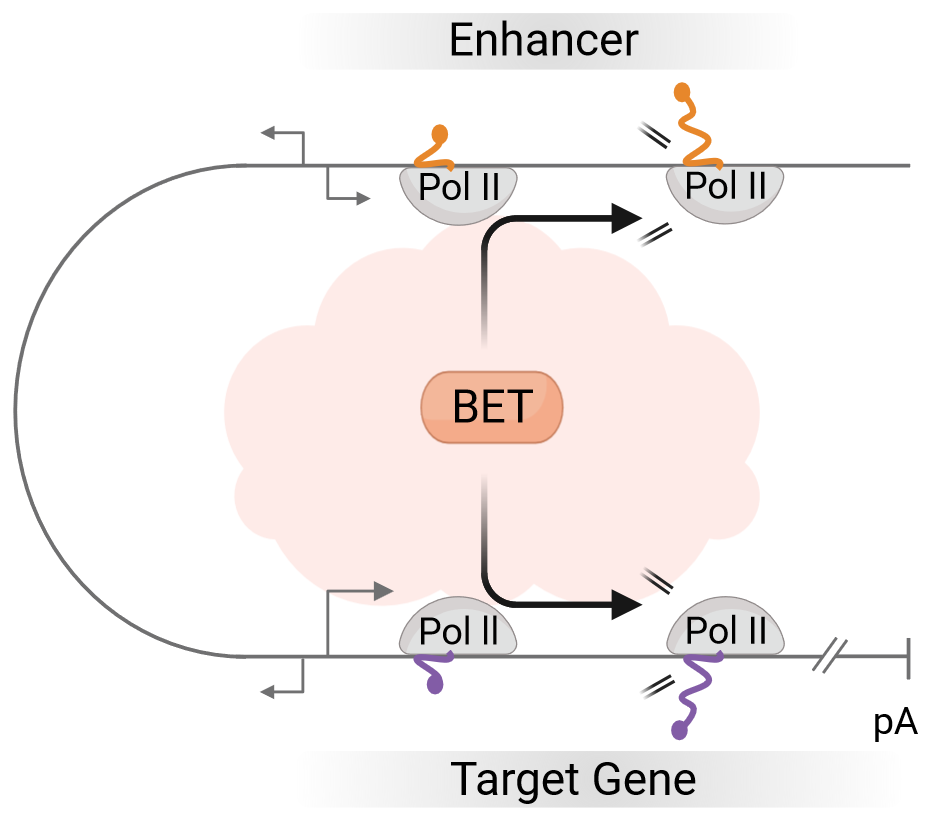
Our knowledge of the regulatory crosstalk between enhancers and their target genes to control the RNA output of genes and cell function is still incomplete. In this collaborative project, we will focus on elucidating the specific role of BET bromodomain proteins in enhancer-target gene communication in their natural environments of the nucleus of human cells. We have recently shown that the most prominent member of the BET protein family, BRD4, is essential for productive nascent RNA synthesis by RNA polymerase II at the majority of genes in human cells, and for coupling transcription with co-transcriptional RNA processing (Arnold, Bressin et al., Mol Cell, 2021; Eischer et al., WIREs RNA, 2023; Altendorfer et al., Transcription, 2022). We recently also found that BRD4 co-localizes to promoter-proximal regions of genes and their enhancers genome-wide, and that acute loss of BRD4 leads to a synchronous collapse of transcription at enhancers and their genes (Bressin, Jasnovidova, Arnold et al., Nature Commun, 2023). Therefore, BRD4 and potentially also other BET proteins are prime candidates to mediate the enhancer-target gene crosstalk. Given the implication of BRD4 in a range of human diseases, such as different types of leukemia, we also plan to extend our studies to selected disease models.
In this project, we will combine functional genomics approaches (CRISPR-Cas9, PROTACs, nascent RNA-seq methods and other techniques) with novel Expansion Microscopy (ExM) approaches to super-resolution microscopy (Gao et al., ACS Nano 2018; Thielhorn et al., Angewandte Chemie Intl. Ed., 2023) and single molecule microscopy methods that are suited to investigate nuclear structures (Geertsema, Nature Biotech, 2021) and especially the partitioning into and out-of condensates (Hoffmann, Rentsch et al., Nature Comms, in press).
For more information on our research programs please see our most recent publications and our lab websites:
© Andreas Mayer
Link to selected recent articles:
Link to a more general description of our research:
Link to websites of the Ewers and Mayer labs:
Membrane Biochemistry - Prof. Dr. Helge Ewers
Max Planck Research Group High-Resolution Functional Genomics, Dr. Andreas Mayer



