Molecular mechanisms of bacterial immunity
Prof. Markus C. Wahl, PhD
Bacteria and viruses that infect them (bacteriophages) wage a constant evolutionary battle. Phages have evolved diverse molecular machinery to attack their hosts and take over host metabolism to ensure their replication. Bacteria, in turn, have acquired tools to fend off or terminate such attacks. The study of phage infection and replication mechanisms can inspire and inform the development of new antimicrobial strategies, while the identification and characterization of bacterial phage defense systems can lead to the development of powerful molecular biological, genetics and therapeutic tools, as impressively exemplified by the restriction/modification and CRISPR/Cas systems, for instance.
Our group is interested in the molecular mechanisms underlying both phage attack and bacterial phage defense systems, with a focus on molecular systems that modulate phage or host gene expression (Fig. 1)1-3.
Numerous novel putative phage defense systems have been identified and in part validated in recent years4-6. However, the detailed molecular mechanisms whereby these systems take effect remain largely unknown. The proposed project will contribute to closing this knowledge gap and will involve:
- Validation of putative phage defense systems of interest (expression of putative phage defense systems in model bacteria; phage infection assays)
- Assessment of cellular protein and nucleic acid interactomes of phage defense system components (in-cell cross-linking in combination with protein mass spectrometry; cross-linking/immunoprecipitation in combination with RNA sequencing; chromatin immunoprecipitation in combination with DNA sequencing)
- Recombinant production of phage defense system components and interactors; reconstitution of their functional complexes (molecular cloning, protein production and purification)
- Characterization of putative enzymatic activities of phage defense system components (in vitro functional assays)
- Structural analysis of phage defense system components, their interactors and their functional complexes (cryogenic electron microscopy in combination with single-particle analysis; macromolecular crystallography; in vitro cross-linking in combination with protein mass spectrometry)
For more information have a look at the website of the Structural Biochemistry group.
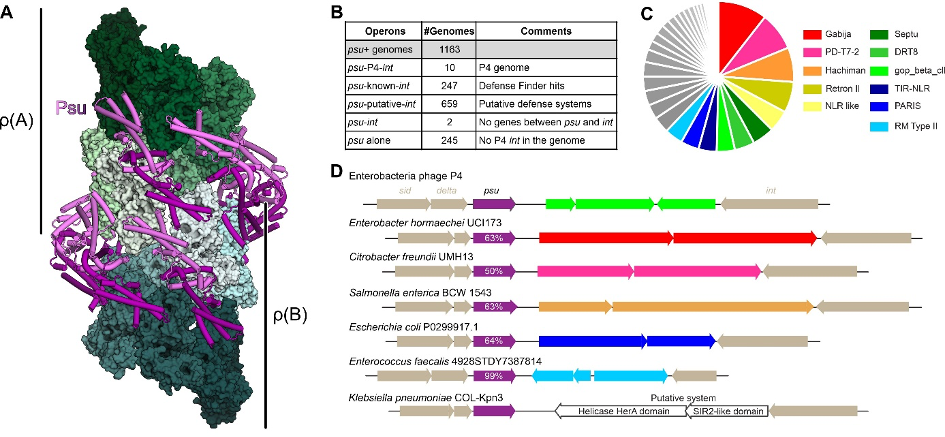
Figure 1. Psu as a marker of anti-phage defense systems. A. Structure of a ρ-Psu complex. Phage P4 capsid protein, Psu, inactivates transcription termination factor, ρ, by forced hyper-oligomerization. Eight Psu dimers (violet and purple) bridge two open ρ rings, ρ(A) and ρ(B), and facilitate expansion of the ρ rings to at least the nonamer stage. B. Defense system analysis of psu+ genomes. C. Psu-like proteins seem to regulate expression of diverse phage defense systems across bacteria. The 37 classes of psu-associated defense systems identified by DefenseFinder4 are shown in a pie chart, with the eleven most abundant defense systems indicated in color. D. Examples of psu-associated defense system loci; % identity to P4 Psu is shown. The known defense genes are colored as in C; a putative defense system with a SIR2-like domain is shown in white. For details see 1.
References