Light microscopy
Our service group hosts a broad range of light microscopes which are operated as shared equipment and are open for all members of the institute. This includes conventional widefield epifluorescence microscopes, automated screening microscopes, confocal laser scanning microscopes as well as a dedicated STED super-resolution microscope, and a 3D light sheet microscope. Furthermore, we support additional instruments located in individual departments and other equipment such as basic cell culture and stereo microscopes, which can be accessed by all users upon request.
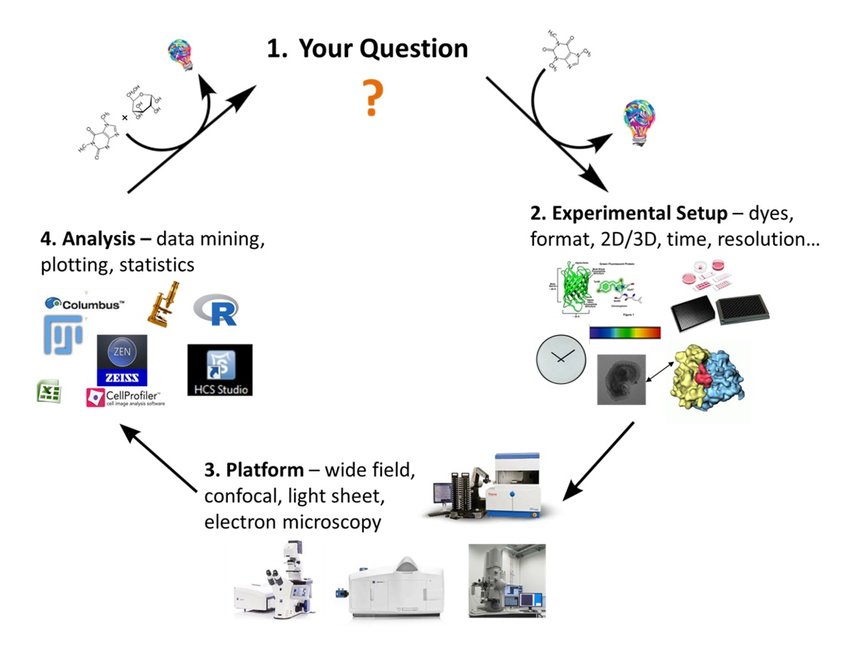
The mission of our group is supporting all users in performing their imaging experiments, might it be a simple routine task or the implementation of a complex and sophisticated imaging and data analysis workflow. In fact, rather than offering standard technical service we think of service as a highly interactive process for developing and supporting a full imaging workflow that involves planning, sample preparation, user training, data acquisition as well as image analysis up to data visualization, statistical analysis and data interpretation (Figure 1).
Frankly speaking, there is no universal imaging procedure available. A simple widefield experiment might be fast and sufficient to detect a certain signal, but might not provide enough resolution to localize it unambiguously. Confocal microscopy might provide higher resolution, but the high laser intensity might bleach the sample and prevent long-term live cell imaging. In particular, imaging large three-dimensional samples requires careful balancing sensitivity, light intensity, resolution, and speed, but also elaborate optimization of sample preparation including sample staining and clearing. Once experiments are up and running, we are open to discuss the results to further optimize all individual steps including sample preparation, data acquisition, image processing as well as data analysis and interpretation.
In addition to our microscopes, we provide access to powerful imaging workstations supporting a wide range of software packages such as Image J, FIJI, Zen Intellesis, Arivis Vision 4D, and Cell Profiler, amongst others. By combining image acquisition and data analysis tools, we develop imaging pipelines based on the specific needs of individual users. Once established, these workflows are then made available to a broader user community. For example, we established analysis pipelines for quantifying RNA- and DNA-fish experiments (collaboration with Melissa Bothe, Bothe et al., 2021, Life Sci Alliance) as well as for quantifying phase separation events accompanying gene regulation inside the nucleus (collaboration with AG Hnisz; Basu et al., 2020, Cell; Asimi et al. ,2022, Nat Genet).
Another example are semi-automated workflows to screen and analyze 3D cell culture systems such as cancer sphaeroids (collaboration with the Yaspo lab and Zeiss) or gastruloids and trunk like structures that are now routinely used to study stem cell differentiation or embryogenesis (Bolondi et al., 2021, Bio Protoc; Veenvliet et al., 2020, Science; Rosebrock et al., 2022, Nat Cell Biol). In short, these experiments are performed in multiwell formats, and time-series are recorded automatically using various autofocus strategies. AI-based segmentation methods are then applied to identify objects of interest, whereas fluorescence images are used for segmentation and quantification of individual cell populations. Similar strategies for automated imaging and data analysis have been implemented to identify e.g. cell cycle intermediates or different cell populations from large 2D or 3D data sets comprising information on thousands of individual cells.