Cryo-electron microscopy and single particle analysis
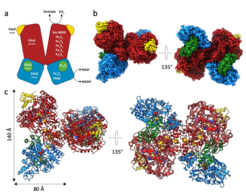
Cryo-EM in combination with the single particle approach has emerged as a key technology in modern structural biology illustrated by the Nobel prize award to three of its pioneers - Jacques Dubochet, Joachim Frank and Richard Henderson - in 2017. In cryo-EM, biological samples are embedded in vitrified ice and then imaged under cryo-low-dose conditions. The introduction of Direct Electron Detector systems (DEDs) provoked a resolution revolution in single particle cryo-EM, which now almost routinely enables near-atomic resolution (Figure 1, Ignatiou et al., 2019, Nat Commun).
Starting already in 2004, we established a state-of-the-art cryo-electron microscopy (cryo-EM) facility within the Berlin-Brandenburg research consortium “UltraStructure Network” (USN), which was at first initiated in close collaboration with Prof. Christian Spahn (Charité Berlin) and Prof. Roland Beckmann (Charité Berlin, now LMU Munich). Our facility provides a technology platform for sample screening and vitrification, automated data acquisition as well as computing resources for image processing. Core instrument of our facility is a 300 kV Tecnai G2 Polara cryo-electron microscope (FEI) equipped with Gatan k2summit direct electron detector. Our group supports all users in the initiation of new cryo-EM projects and can provide access to two even more powerful Titan Krios cryo-TEMs, which have recently been installed at the FU Berlin and Charité (Berlin Buch), respectively.

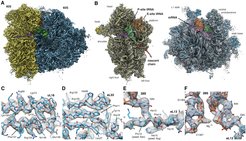
In single particle cryo-EM, 3D information is derived by averaging over thousands if not hundred thousand of projection images of individual molecules (referred to as particle images), assuming that they are all identical. In practice, however, protein complexes typically show variable assembly and intrinsic conformational flexibility which often represent different biological relevant functional states. Within the collaborative research center SFB740 (project Z1), we established a fully automated pipeline for high-throughput data collection including on-the-fly drift correction and CFT-based quality control of DED super-resolution movie stacks which was essential for introducing multi-particle refinement strategies developed in the Spahn lab (Charité). Combining these tools enabled us to study for the very first time ex-vivo derived protein complexes such as actively translating human polysomes derived from HEK cells, which represent heterogeneous samples by nature. Analyzing polysome preparations, we could decipher 11 distinct functional states, 10 of which representing intermediates of the ribosomal elongation cycle and refine the highest-occupied state, the 80S POST-complex, to near-atomic resolution (Figure 2, Behrmann et al., 2015, Cell).
Multi-particle refinement was then applied to study phage assembly intermediates (Ignatiou et al., 2019, Nat Commun), transcription regulation (Huang et al., 2020, Mol Cell; Said et al., 2021, Science), ribosome assembly (Nikolay et al., 2018, Mol Cell; Nikolay et al., 2021, Mol Cell; Quin et al, unpublished data), and translational control in embryonic brain development (Kraushar et al., 2021, Mol Cell). Moreover, the resolution power of DED camera systems now even allows structural analysis of small enzyme complexes comprising a molecular weight of only 200-300 kDa and below such as e.g. the metal-containing formate dehydrogenase (FDH) from Rhodobacter capsulatus (Figure 3, Radon et al., 2020, Nat Commun).