Metabolomics
Metabolites are small molecules participating in metabolic reactions, which are necessary for cellular function, maintenance and growth. Typically, metabolites range from 50 to 1500 Da, while their concentrations span several orders of magnitude. The metabolome is highly dynamic, time-dependent, and metabolites are sensitive to many environmental conditions. It is still unknown how many different metabolite species exist within a cell or organism; predicted are at least 2000 in mammals. Metabolites are extensively exchanged with the environment, e.g., food intake, excretion, inhalation, secondary metabolites, such as medications, flavorings, and recreational drugs, which can be further processed by the gut microbiome or organs. Furthermore, metabolites are, chemically, very diverse (polarity, charge, pKa, solubility, volatility, stability, and reactivity), consequently no single method can capture and analyze the entire metabolome at once.
In our lab, we established a targeted LC-MS approach, a high-quality analysis of a subset of predefined metabolites with known chemical structures. Once established, the target-based approach is advantageous because of its reliable measurement of the selected analytes in complex samples. Multiple reaction monitoring (MRM) has been commonly used for the analysis of small molecules and peptides, but the identification of metabolites based on one MRM, is still error-prone. As an additional layer of confidence in metabolite identification, we employ metabolite-specifictransition ratios, MRM ion ratios.
Thus, the combination of three MRMs and their MRM ion ratios results in an enhanced method for the reliable, rapid, and quantitative profiling of metabolites.
Multiple reaction monitoring (MRM)
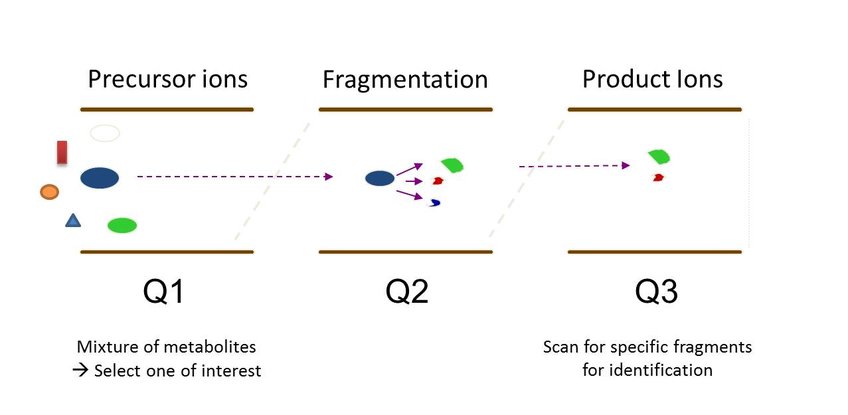
A complex mixture of metabolites can be used as starting material. The parent mass of a metabolite of interest is selected in quadropole 1 (Q1) of the mass spectrometer (Figure 1) and sent to Q2 for fragmentation. Basicly, the metabolite will be fragmented during colliding with a collission gas in Q2. In Q3, fragments will be isolated and sent to the detector. Resulting fragments are usually specific for one metabolite. Fragment pattern can be compared to own standards or databases.
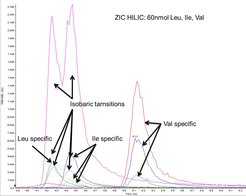
Isobaric metabolites, like leucine and isoleucine have to elute from analytical columns at different times in order to distinguish them (Figure 2).
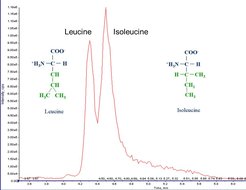
As all amino acids share the same basic structure, they can only be distinguished by their side chains or different elution times. Figure 3 features the branched chain amino acids; isobaric amino acids leucine and isoleucine elute at slightly different retention times. Some transitions are shared between them, as these fragments originate from the basic structure; some transitions are specific, because the fragments contain at least unique parts of the side chain. Thus, different elution times are not always necessary. Unfortunately, the intensity of specific fragments is sometimes very low.