Proteomics
The proteome is the entire set of proteins expressed by a cell/organism at a certain time. The goal of proteomics is a comprehensive, quantitative description of protein expression and its changes under the influence of biological perturbations (Anderson and Anderson, Electrophoresis, 1998). Liquid chromatography mass spectrometry-based (LC-MS) proteomics allow a large-scale identification and quantification of proteins from complex samples. Several thousands of proteins can be analyzed in a single LC-MS run.
Q Exactive HF OrbiTrap
A general proteomic and phospho-proteomic profiling scheme, which we use in our lab is shown in Figure 1.
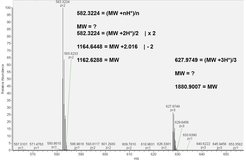
Here, you see MS OrbiTrap spectra of two different peptides. Multiple peaks per peptide are caused by 13C6 isotopes. How can you calculate the molecular weight (MW) of a peptide?
Peptides need to be further fragmented in order to read the sequence. There are different fragmentation methods available, resulting in different fragmentation patterns. The Q Exactive HF is applaying Higher-energy C-trap dissociation (HCD), specific y and b ions are generated as shown in Figure 3.
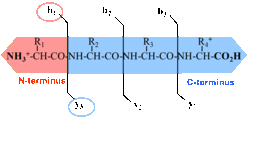
Figure 3: Peptide fragment ions are indicated by b, if the charge is retained on the N-terminus and by y, if the charge is maintained on the C-terminus
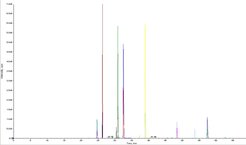
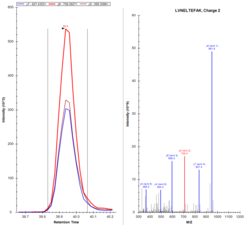
QTrap 6500, targeted MRM analysis
For the QTrap 6500, peptide specific methods have to be established and optimized. At least three transitions per peptide are necessary for identification, they can be quantitated and compared to other samples. Figure 4 shows several transitions from our BSA standard run over 75 minutes, Figure 5 displays three MRM transition from one specific peptide in detail and its according MS2 spectrum.