Understanding the Distribution of Mutations along Genomes
Dr. Peter Arndt
Keywords: population genetics, coalescent theory, cancer genomics
The genetic information stored in DNA is subject to a variety of mutagenic processes. These processes can alter a single base pair, resulting in a single nucleotide polymorphism (SNP). Alternatively, they can insert or delete a stretch of sequence (indels). More complex changes may also occur, such as inversions or ectopic rearrangements. It is crucial to gain a comprehensive understanding of the specific distribution of the effects of these processes along the DNA. Local patterns will influence not only evolutionary processes but also the development of diseases such as cancer.
Recent advances in efforts to sequence the whole genome of many individuals in a population and the availability of these data give us the opportunity to reevaluate and extend models of DNA mutation. Our modeling will also incorporate the demographic history of populations. In accordance with coalescent theory, this approach will facilitate the elucidation of intricate patterns of DNA alteration along chromosomes.
Successful candidates will be responsible for developing projects designed to study sequencing data from human (or other species) population samples, in addition to sequencing data from cancer samples. A robust background in dynamical modeling, population genetics, coalescent theory, statistical analysis, computational biology, and machine learning is essential. Prior experience with the Julia programming language is an additional advantage.
For more information visit the website of the Evolutionary Genomics Group.
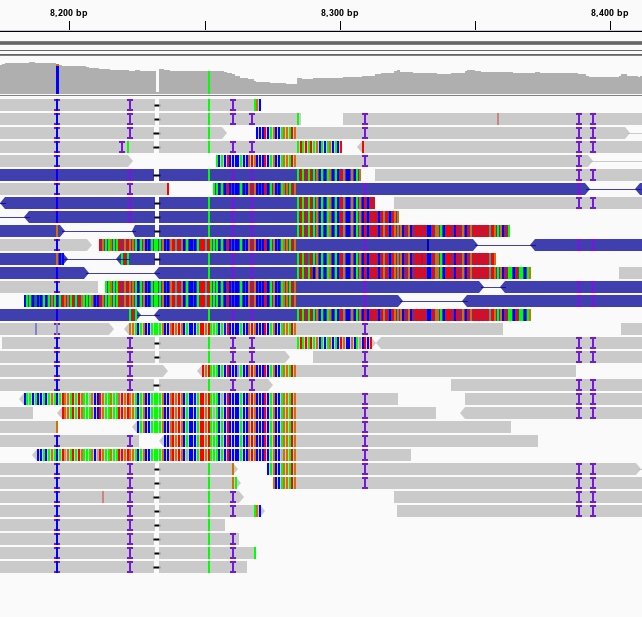
Sequencing reads aligned to the human reference genome as seen in the Integrative Genomics Viewer (IGV). Missmatches and Indels are highlighted using different colors.


