Computational systems medicine and disease control
Prof. Dr. Max von Kleist
Our group develops methods for computational systems medicine and computational systems biology and applies them to questions arising in fundamental science and infectious disease research. We welcome students with a solid background in mathematical biology (data science, modelling and simulation) and theoretical foundations in bioinformatics who are excited about the research in these areas. Specifically, we are looking for a highly qualified student to work on:
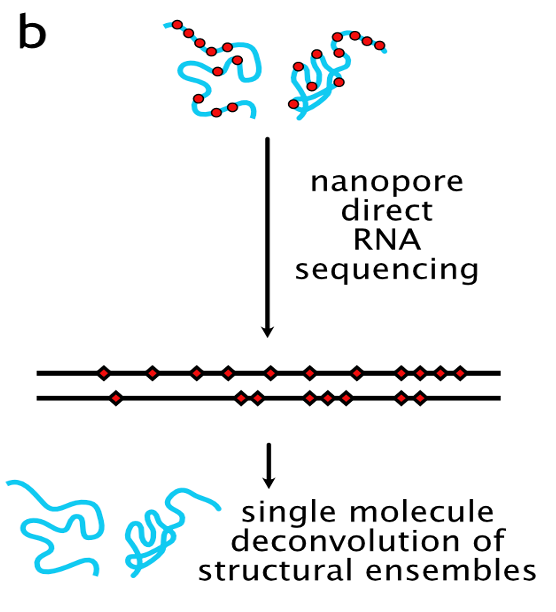
1. Learning RNA structural ensembles from direct (nanopore) RNA sequencing of chemically probed molecules:
RNA is a functionally diverse molecule that is a central player in the regulation of many biological processes where they execute a vast array of cellular functions. As such, RNA is increasingly recognized as a therapeutic target, therapeutic drug, and as a tool for synthetic biology. RNA function is based on its capacity to fold into intricate 3d structures formed from regions of single-stranded and double-stranded RNA. Axiomatic to RNA’s functional plasticity is structural heterogeneity. Rather than folding into a single structural conformation, RNA molecules can often adopt several different folds, each with a different function.
Experimentally, structural information can be derived by ‘chemical probing’1 of RNA molecules. The chemical probes selectively modify RNA residues that are unpaired (‘free’ Watson-Crick edge). Following a ‘probing experiment’, chemical modifications can be detected by next-generation sequencing, mapped onto the sequence and used for predicting RNA structure(s). Within the domain of RNA structure-function inference, we intend to further our existing technical expertise1,2,3. In particular, we intend to develop computational (ML) methods to resolve RNA structural ensembles of the HIV-1 transcriptome in cells from high-throughput data provided by our experimental partners at the HIRI Würzburg that combine chemical probing1 with long-read nanopore direct RNA sequencing (dRNA-seq)5. A particular challenge will be the differentiation between sequencing- and base-calling errors during direct RNA sequencing6, as well as multiplex dRNA sequencing to reduce inter-experimental noise and reduce costs7. The project will involve aspects of bioinformatics, data science & statistics, as well as supervised and unsupervised machine learning. In particular, training of recurrent neural networks for nanopore base- and modification calling, as well as unsupervised methods for cluster detection (= structural conformations) in base-called read-ensembles will be the key to this project.
2. Modelling of HIV-1 chemoprevention and SARS-CoV-2 evolution
We are interested in modelling the efficacy of pharmaceutical interventions in preventing viral infection (HIV-1), as well as in modelling the interplay between immunity and viral evolution (SARS-CoV-2).
We hypothesize that the evolution of SARS-CoV-2 is largely driven by population immunity and the ability to cross-neutralize incident variants. To study this phenomenon, we are combining deep mutational scanning data8 with (molecular) surveillance data9 and infection history10 to quantify, for each emerging strain, the strain-specific expected number of susceptible individuals11.
Quantifying the clinical efficacy of preventive therapies (vaccine, prophylaxis) poses an immense statistical (and monetary) challenge: As an example, a single COVID vaccine had to be tested on 40,000 individuals to obtain ~100 evaluable data points (= infections) distributed over two intervention arms (placebo, e.g. 95 infections vs. vaccine, 5 infections). To allow investigating dose-effect relationships and to derive a mechanistic understanding of the mode-of-action of preventive measures, we develop integrative stochastic modelling and simulation techniques in the field of systems medicine and systems pharmacology12,13. We use these models14 in collaboration with clinical- and epidemiological partners at RKI, Johns Hopkins, Harvard to consult decision makers and to develop guidelines with the WHO.
These projects require a good understanding of mathematical modelling and stochastic simulation techniques12,13.
References:
1General Principles for the Detection of Modified Nucleotides in RNA by Specific Reagents. M. Helm, …, Y. Motorin. Advanced Biology, 5, 2100866
2Mutational Interference Mapping Experiment (MIME) for studying the relationship between RNA structure and function. R.P. Smyth*, L. Despons, G. Huili, S. Bernacchi, M. Hijnen, J. Mak, F. Jossinet, L. Weixi, J-C. Paillart, M. von Kleist*, R. Marquet*, Nature Methods, 12, 866 , 2015
3In cell Mutational Interference Mapping Experiment (in cell MIME) identifies 5’ PolyA as a dual regulator of HIV-1 genomic RNA production and packaging, R.P. Smyth*$, M.R. Smith$, A-C. Jousset, L. Despons, G. Laumond, T. Decoville, P. Cattenoz, C. Moog, F. Jossinet, M. Mougel, J.-C. Paillart, M. von Kleist*, R. Marquet*, Nucleic Acids Research, 46, e57, 2018
4Short and long-range interactions in the HIV-1 5'UTR regulate genome dimerization and Pr55Gag binding, L. Ye, ..., M. von Kleist, R.P. Smyth, Nature Struct & Mol Biol, 29, 306 2022
5Computational methods for RNA modification detection from nanopore direct RNA sequencing data. M. Furlan, …, T. Leonardi. RNA Biology, 18, 31, 2021
6Sequencing accuracy and systematic errors of nanopore direct RNA sequencing. W. Liu-Wei, W. Van der Toorn, P. Bohn, M. Hölzer, RP. Smyth, M. von Kleist. BioRXiV https://doi.org/10.1101/2023.03.29.534691
7Demultiplexing and barcode-specific adaptive sampling for nanopore direct RNA sequencing. W. van der Toorn* , P. Bohn*, W. Liu-Wei , M. Olguin-Nava, RP Smyth+, M. von Kleist + , in review (2024).
8 Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Cao, Y., Wang, J., Jian, F. et al., Nature 602, 657–663 (2022).
9 Integrated Molecular Surveillance for SARS-CoV-2 (IMS-SC2) Laboratory Network. Advancing Precision Vaccinology by Molecular and Genomic Surveillance of Severe Acute Respiratory Syndrome Coronavirus 2 in Germany, 2021. Oh DY, Hölzer M, …, Kröger S*, von Kleist M*, Wolff T*, Clin Infect Dis. 2022 Aug 15;75(Suppl 1):S110-S120
10 Rapid incidence estimation from SARS-CoV-2 genomes reveals decreased case detection in Europe during summer 2020. Smith, M.R., Trofimova, M., …, von Kleist M., Nat Commun 12, 6009 (2021)
11SARS-CoV-2 Evolution on a Dynamic Immune Landscape. Raharinirina N. A. *, Gubela N.*, Börnigen D.*, Smith M. R.*, Oh D.-Y., Budt M., Mache C., Schillings C., Fuchs S., Dürrwald R., Wolff T., Hölzer M., Paraskevopoulou S., von Kleist M. in review, 2023.
12Hybrid stochastic framework predicts efficacy of prophylaxis against HIV: An example with different dolutegravir regimen, S. Duwal, L. Dickinson, S. Khoo and M. von Kleist, PLoS Computational Biology, 14, e1006155, 2018
13Numerical approaches for the rapid analysis of prophylactic efficacy against HIV with arbitrary drug-dosing schemes. L. Zhang, J. Wang, M. von Kleist, PLoS Comput Biol 17(12): e1009295
14Model-based predictions of protective HIV PrEP adherence levels in cisgender women. L. Zhang*, S. Iannuzzi*, A. Chaturvedula, E. Urungu, J. Haberer, CH Hendrix, M. Von Kleist, Nature Medicine, 2023; 29: 2753.
For more information visit the website of the Systems Pharmacology group.
© M. von Kleist