TADs in Evolution
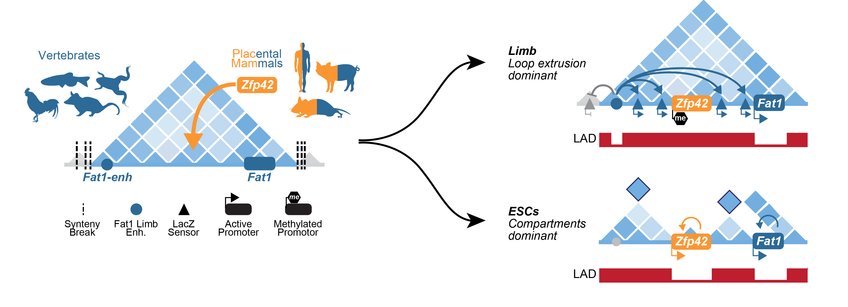
TADs are relatively stable during evolution and their disruption can lead to gene misexpression and disease. Modifying such regulatory landscapes and their transcriptional outputs is viewed as central for acquiring novel phenotypic traits in evolution. But how are regulatory landscapes modified to incorporate novel activities without compromising their existing functions? Though TADs transmit enhancer activities to all positions in a domain, newly emerged or reshuffled genes do not universally adopt all these regulatory inputs. Indeed, many TADs generated by evolution contain multiple genes with non-overlapping expression despite all promoters contacting the same enhancers. Evolutionary altered regulatory landscapes must therefore employ additional mechanisms that further refine how and when promoters use enhancer activities.
In this project we address this question by reconstructing how a new gene regulatory program could emerge during evolution within a more ancient TAD without disrupting its prior activities. We investigate how the Zfp42 gene emerged within the ancient Fat1 TAD in placental mammals. We find that multiple novel expression programs can be incorporated into a single locus during evolution by at least restructuring 3D-chromatin landscapes and selective promoter silencing. In this way, we demonstrate generalizable principles of how the genome resolves regulatory conflicts that inevitably arise in development and evolution.