4D Genome Architecture, Gene Regulation & Developmental Disease
Most developmentally important genes have complex and pleiotropic expression patterns. Such spatially and temporally restricted transcriptional activities are largely controlled by cis-regulatory elements such as enhancers that can be far away from their target gene. Our current knowledge how enhancers activate their target genes is limited. How genes are regulated and how this works within the nuclear space remains elusive and is probably some of the biggest challenges in the post-genomic era.
The folding of chromosomes in the nucleus is tightly controlled. This folding process has a dual role, it compacts the genomic information within the nucleus and, at the same time, has an important role in gene regulation. Studies using technologies such as HiC have shown that the genome is partitioned into megabase scale compartments called topologically associated domains (TADs). Because TADs restrict the range of enhancer targets, enhancers usually contact genes located within these TADs, but not outside. TADs have been shown to be surprisingly stable across cells, tissues and even species, suggesting that they function as a general folding scaffold determining domains of possible interaction partners. We previously showed that deletions, duplications, inversions, translocations and insertions, collectively called structural variations (SVs), can result in disease-causing rewiring of enhancer-promoter contacts by changing the TAD configuration at a locus.
Using primarily mouse limb development as a model system, we investigate how changes in 3D genome organization impact on gene regulation during development and disease. We are interested in fundamental principles of enhancer-mediated gene regulation and the rewiring of 3D architecture by SVs. In addition, we investigate the role of long-non-coding RNAs and transposable elements in gene regulation, 3D genome organisation and developmental disorders. To accomplish this, we are using a large variety of state-of-the-art technologies: on the one hand, we employ Chromosomal Conformation Capture technologies (4C, Hi-C), ChIP-seq, and RNA-seq to describe regulatory landscapes. On the other hand, we apply CRISPR-Cas9 genome editing to induce various genetic re-arrangements (deletions, inversions, duplications, insertions) and study their impact on chromatin architecture, gene regulation, and limb development.
Publications
1.
Lila Allou, Sara Balzano, Andreas Magg, Mathieu Quinodoz, Beryl Royer-Bertrand, Robert Schöpflin, Wing-Lee Chan, Carlos E. Speck-Martins, Daniel Rocha Carvalho, Luciano Farage, Charles Marques Lourenço, Regina Albuquerque, Srilakshmi Rajagopal, Sheela Nampoothiri, Belinda Campos-Xavier, Carole Chiesa, Florence Niel-Bütschi, Lars Wittler, Bernd Timmermann, Malte Spielmann, Michael I. Robson, Alessa Ringel, Verena Heinrich, Giulia Cova, Guillaume Andrey, Cesar A. Prada-Medina, Rosanna Pescini-Gobert, Sheila Unger, Luisa Bonafé, Phillip Grote Carlo Rivolta Stefan Mundlos✉ & Andrea Superti-Furga
Non-coding deletions identify Maenli lncRNA as a limb-specific En1 regulator
Nature (2021)
2.
Konstantin Helmsauer*, Maria E. Valieva*, Salaheddine Ali*, Rocío Chamorro González,
Robert Schöpflin, Claudia Röefzaad, Yi Bei, Heathcliff Dorado Garcia, Elias Rodriguez-Fos,
Montserrat Puiggròs, Katharina Kasack, Kerstin Haase, Csilla Keskeny, Celine Y. Chen,
Luis P. Kuschel, Philipp Euskirchen, Verena Heinrich, Michael I. Robson, Carolina Rosswog,
Joern Toedling, Annabell Szymansky, Falk Hertwig, Matthias Fischer, David Torrents,
Angelika Eggert, Johannes H. Schulte, Stefan Mundlos*, Anton G. Henssen*, &
Richard P. Koche*
Enhancer hijacking determines extrachromosomal circular MYCN amplicon architecture in neuroblastoma
Nat Commun 11, 5823 (2020)
3.
Christina Palioua, Philine Guckelbergera, Robert Schöpflina, Verena Heinrichd, Andrea Espositoe,
Andrea M. Chiarielloe, Simona Biancoe, Carlo Annunziatellae, Johannes Helmuthh, Stefan Haasd, Ivana Jerkovi�ca,
Norbert Brieskea, Lars Wittleri, Bernd Timmermannj, Mario Nicodemie, Martin Vingrond, Stefan Mundlos,
and Guillaume Andrey
Preformed chromatin topology assists transcriptional robustness of Shh during limb development
Proceedings of the National Academy of Sciences Jun 2019, 116 (25) 12390-12399
4.
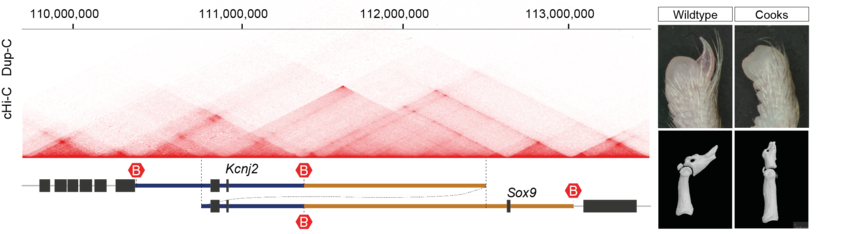
Alexandra Despang, Robert Schöpflin, Martin Franke, Salaheddine Ali, Ivana Jerković,
Christina Paliou, Wing-Lee Chan, Bernd Timmermann, Lars Wittler, Martin Vingron,
Stefan Mundlos* and Daniel M. Ibrahim*
Functional dissection of the Sox9–Kcnj2 locus identifies nonessential and instructive roles of TAD architecture
Nat Genet 51, 1263–1271 (2019).
5.
Kragesteen BK, Spielmann M, Paliou C, Heinrich V, Schöpflin R, Esposito A, Annunziatella C, Bianco S, Chiariello AM, Jerković I, Harabula I, Guckelberger P, Pechstein M, Wittler L, Chan WL, Franke M, Lupiáñez DG, Kraft K, Timmermann B, Vingron M, Visel A, Nicodemi M, Mundlos S, Andrey G.
Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis.
Nat Genet. 2018 Oct;50(10):1463-1473. Epub 2018 Sep 27.
6.
Spielmann M, Lupiáñez DG, Mundlos S.
Structural variation in the 3D genome.
Nat Rev Genet. 2018 Jul;19(7):453-467. Review.
7.
Bianco S, Lupiáñez DG, Chiariello AM, Annunziatella C, Kraft K, Schöpflin R, Wittler L, Andrey G, Vingron M, Pombo A, Mundlos S, Nicodemi M.
Polymer Physics Predicts the Effects of Structural Variants on Chromatin Architecture
Nat Genet. 2018 May;50(5):662-667. Epub 2018 Apr 16.
8.
Andrey G, Mundlos S.
The three-dimensional genome: regulating gene expression during pluripotency and development.
Development. 2017 Oct 15;144(20):3646-3658. Review.
9.
Will AJ, Cova G, Osterwalder M, Chan W-L, Wittler L, Brieske N, Heinrich V, de Villartay J-P, Vingron M, Klopocki E, Visel A, Lupiáñez DG, Mundlos S.
Composition and dosage of a multipartite enhancer cluster control developmental expression of Ihh (Indian hedgehog).
Nat Genet. 2017 Oct;49(10):1539-1545. Epub 2017 Aug 28.
10.
Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, Kraft K, Kempfer R, Jerković I, Chan WL, Spielmann M, Timmermann B, Wittler L, Kurth I, Cambiaso P, Zuffardi O, Houge G, Lambie L, Brancati F, Pombo A, Vingron M, Spitz F, Mundlos S.
Formation of new chromatin domains determines pathogenicity of genomic duplications.
Nature 2016 Oct 13;538(7624):265-269. Epub 2016 Oct 5.
11.
Lupiáñez DG, Spielmann M, Mundlos S.
Breaking TADs: How Alterations of Chromatin Domains Result in Disease.
Trends Genet. 2016 Apr;32(4):225-237. Epub 2016 Feb 7.
12.
Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, Santos-Simarro F, Gilbert-Dussardier B, Wittler L, Borschiwer M, Haas SA, Osterwalder M, Franke M, Timmermann B, Hecht J, Spielmann M, Visel A, Mundlos S.
Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions.
Cell. 2015 May 21;161(5):1012-1025. Epub 2015 May 7.
13.
Kraft K, Geuer S, Will AJ, Chan WL, Paliou C, Borschiwer M, Harabula I, Wittler L, Franke M, Ibrahim DM, Kragesteen BK, Spielmann M, Mundlos S, Lupiáñez DG, Andrey G.
Deletions, Inversions, Duplications: Engineering of Structural Variants using CRISPR/Cas in Mice.
Cell Rep. 2015 Feb 4. pii: S2211-1247(15)00029-7. [Epub ahead of print]
14.
Ibn-Salem J, Köhler S, Love MI, Chung HR, Huang N, Hurles ME, Haendel M, Washington NL, Smedley D, Mungall CJ, Lewis SE, Ott CE, Bauer S, Schofield PN, Mundlos S, Spielmann M, Robinson PN.
Deletions of chromosomal regulatory boundaries are associated with congenital disease.
Genome Biol. 2014 Sep 4;15(9):423.