Genome structure
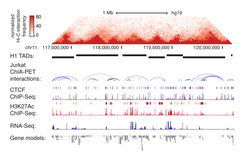
Understanding how transcriptional enhancers control over 20,000 protein-coding genes to maintain cell-type-specific gene expression programs in all human cells is a fundamental challenge. Recent studies suggest that gene regulatory elements and their target genes generally occur within insulated neighborhoods, which are chromosomal loops formed by the interaction of two DNA sites bound by the CTCF protein and occupied by the cohesin complex. Insulated neighborhoods provide for specific enhancer-gene interactions, are essential for both normal gene activation and repression, are largely preserved throughout development, form the mechanistic basis of higher order genome folding (e.g. Topologically Associating Domains) and are perturbed by genetic and epigenetic factors in disease.
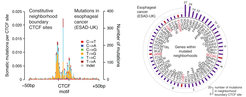
We develop novel assays to identify protein components involved in the control of insulated neighborhoods. We develop state-of-the-art genetic and protein perturbation approaches to dissect regulatory pathways of insulated neighborhood function. We ascertain mutations that alter neighborhood anchors in cancer cells, and explore models how such mutations promote oncogenic expression programs.
Further reading
Dowen et al., Cell 2014
Hnisz et al., Science 2016
Hnisz et al., Cell 2016
Hnisz et al., ARCB 2017

