Evolutionary Genomics of Limb Adaptation
Evolution has created an astonishing array of morphological diversity. A striking example is the adaptation of limbs during evolution. The transformation of fins into limbs was fundamental to the transition from aquatic to terrestriallife. However, since then, the tetrapod limb has been subject to extensive evolutionary adaptation to generatefunctional diversity. Darwin highlighted the differences in limb morphology among living creatures to support hisargument for evolution by natural selection and descent in the often quoted statement “what can be more curiousthan that the hand of a man, formed for grasping, that of a mole for digging, the leg of the horse, the paddle of theporpoise, and the wing of the bat, should all be constructed on the same pattern, and should include the same bones, in the same relative positions?” (The origin of species; 1859).
This project focuses on how such diversity emerges and is codified in the genome. We compare genomic sequences from different species and search for changes that are related to morphological diversity. Based on our knowledge about mutational mechanisms in human disease, we hypothesize that similar processes can lead to evolutionary changes, neofunctionalization and the development of novel traits. In the past years, we have extensively shown that structural variations (deletions, duplications, inversions) can alter gene regulatory landscapes, thereby resulting in ectopic enhancer-promoter contacts and gene misexpression. In an evolutionary context, the rewiring of regulatory information can confer new expression domains and new functions to a gene without changing the specificities of the protein itself. Such changes can have drastic effects and are thus prime candidates for natural selection.
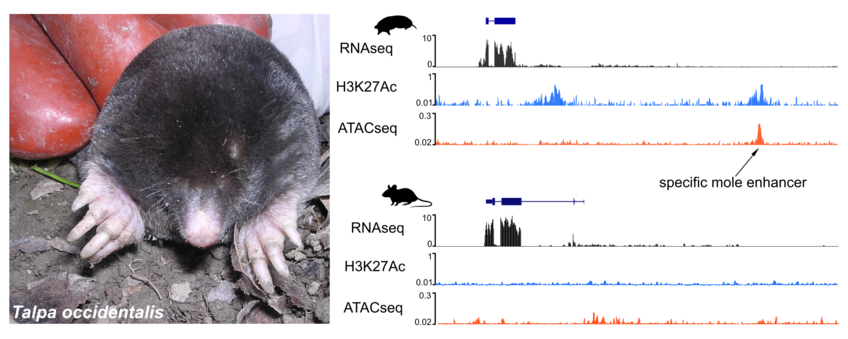
We use the Iberian mole, Talpa occidentalis, as a model of study because it presents very specific and unique traits, including high tolerance to low O2 levels, very small hypoplastic eyes, forelimb polydactyly and the presence of testis-like tissue in females. In fact, moles are the only case of true hermaphroditism in mammals (females develop testes and ovaries (ovotestis)).
In our second project, we focus on an extreme example of evolutionary adaption, the development of wings in bats (Chiroptera). With their forelimbs adapted as wings, they represent the only known example of flight in mammals. Unlike most quadrupedal animals where the differences between fore- and hindlimbs are relatively small, the bat forelimb has developed into a wing and thus differs dramatically from the hindlimb. The batwing is made of a membrane supported by the upper arm (chiropatagium) and the greatly elongated metacarpals and digits II-V of the hand, which support the distal part of the wing. We focus on identifying regulatory regions in the bat genome that govern three major phenotypes: digit elongation, interdigital webbing, and reduction of the ulna, all essential components of the bat limb.
We use genomic approaches, including whole genome sequencing with short and long read technologies and a variety of functional assays (RNAseq, ChIP-seq for histone modifications, ATAC-seq) to investigate genetic changes that occur through evolution and to detect those that might be linked to the phenotype of interest. In a more global context, by comparing non-model species, like moles and bats, with other close-related or more distant organisms, we want to investigate common mechanisms of changes in gene regulation and their contribution to evolution and the acquisition of traits.