A landscape of mammalian development
New technology enables single cell analysis of mouse embryonic development
Scientists from Seattle and Berlin have published an atlas on mouse embryonic development. In their study the researchers examined about two million cells, with the RNA of each cell labeled individually with a specially developed method termed sci-RNA-seq. Junyue Cao, Malte Spielmann and their colleagues describe, which cell types differentiate between days 9.5 and 13.5 of mouse embryonic development, and how they transform into organs. In total, the scientists were able to identify 38 different main cell types and over 500 subtypes. In addition, they described 56 developmental trajectories in organ development for different cell types. The researchers compiled their results in an online atlas and made them accessible to the public.
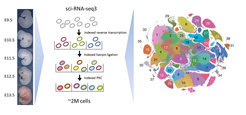
Organ development of mammals is an amazing process. After the fusion of the egg cell with the sperm, the zygote transforms into a cluster of cells by serial cell divisions. Initially, all these cells are identical, but soon, they start to form the three germ layers, which represent the first stage of differentiation of the developing embryo. Within a short time, the cells of the three germ layers are transformed into an embryo containing most of its major internal and external organs.
All of this takes place in the first third of embryonic development. Although all cells contain the same genetic material, they develop into different directions. In order to realize this, different genes have to be activated in each cell at different times. Mistakes during this process, that is, if a wrong gene is active or inactive at the wrong place or at the wrong time, can lead to developmental defects.
Analysis of gene activity
The most common method for investigating embryonic developmental disorders is to concentrate on a single organ system in the mouse and conduct gene knockout studies. For this, individual genes are switched off systematically and their effects on the on the developing organs or whole organisms are examined.
However, this approach, which is mostly performed on mice, is limited to the study of individual animals and tissues. It lacks the necessary throughput and resolution to provide a global overview of the dynamic molecular processes that take place in the different and rapidly growing populations and subpopulations of cells during organogenesis. "Single cell analysis is a promising alternative for such analyses," explains Malte Spielmann, group leader at the Max Planck Institute for Molecular Genetics in Berlin. Spielmann has spent the last two years as postdoc with Jay Shendure and Cole Trapnell at the University of Washington, Seattle, USA, where he worked on the development of a single cell RNA labeling system.
Together with researchers from Berlin, the American team used this method to study the activity of all cells from mouse embryos at the age of 9.5 to 13.5 days. In the current issue of the journal Nature, the scientists describe how the method has been improved and applied to mouse embryos in order to create an "atlas" of gene activity during mouse organogenesis.
Labeling of the transcriptome of single cells
In order to find out which genes of a cell were active at a certain point of development, the scientists examined the transcriptome. This is the set of all RNA molecules of a cell reflecting the genes that are actively expressed at that time. The new method named Single Cell Combinatorial Indexing allows it to tag all RNA molecules of an individual cell with a specific molecular barcode without the need for physical isolation of each cell.
Using this technique, the scientists from Seattle and Berlin have labeled the RNA of all cells from 61 mouse embryos between day 9.5 and day 13.5. Thus, each RNA molecule could be assigned to an individual embryo and its individual stage of development. Subsequently, the RNA of each individual cell was sequenced and analyzed. As a result, the researchers could say exactly which genes of a respective embryo cell were active at the time of the investigation.
Multitude of previously unknown cell types identified
Over 2 million of individual cells have been studied by the scientists. "This way, we were able to show exactly, which genes were involved in the development of the different organs and at what time the major steps of development took place," explains Spielmann. Bioinformatic analysis of this huge amount of data was performed by Junyue Cao. Cao and Spielmann, both first authors of the current study, identified 38 main cell types and more than 500 further granular cell types ("subtypes") - many have never been described before.
Using a software called Monocle 3 that has been developed specifically for this study; the team was also able to demonstrate the dynamics of gene expression within individual cell types during embryonic development. In addition, the relative proliferation rates of cell types and subtypes could be quantified and for the first time, 56 cellular development pathways (trajectories) in organ development could be described.
MOCA – a single cell atlas of mouse organogenesis
The scientists have created a MOCA, a Mouse Organogenesis Cell Atlas to compile their results and make them freely available. “MOCA is an important step in the investigation of embryonic defects and represents a fundamental resource for the developmental biology of mammals”, Spielmann says. ”Single cell analysis of whole embryos enables classical questions of developmental biology to be dealt with digitally. This represents a first step towards the digital embryo and will help to avoid some of the animal testing in the future," he hopes. Unfortunately, animal experiments are still needed for basic research in the field of developmental biology.
The methods for single cell labeling and bioinformatics analysis provide the technical capabilities for individual laboratories for studying single cell. MOCA and all underlying data are freely accessible and can be used by the entire scientific community to contribute to a better understanding of mutations and embryonic malformations.












