Neurodevelopment and translation control
Kinkley Lab
The higher order organization of DNA and chromatin in the nucleus impacts all nuclear processes including transcription, replication, DNA repair and cell division. Yet compaction past the 10nm fiber remains poorly understood. Our lab is interested in elucidating the mechanisms of higher chromatin order, the players involved and how dysregulation of these processes lead to disease or malignant phenotypes.
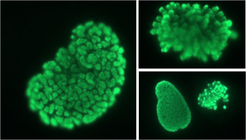
Light microscopy images of U2OS mitotic prophase and metaphase cells (100x magnification). This represents the highest levels of higher chromatin order in human cells.
Lab Projects:
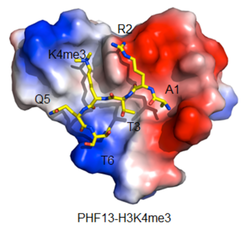
PHF13 Project: PHF13 is an H3K4me3 epigenetic reader involved in higher chromatin order, transcriptional regulation, DNA damage response, cell division and differentiation. We are working towards understanding the molecular interactions of PHF13 to elucidate how it modulates so many fundamental cellular processes and its impact on genome stability.
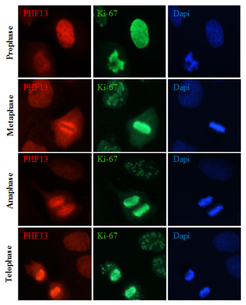
PHF13 and Higher Chromatin Order: PHF13 has been shown to impact mitotic chromatin compaction and structure, indicating a role in the highest levels of chromatin organization. Similarly, it has been shown to interact with several heterochromatin promoting proteins such as HP1 and Polycomb (PRC2), indicating a role as well in heterochromatin formation or maintenance. We strive to better understand the importance and function of PHF13 in these processes.
Selected Publications:
Fuchs A., Torroba M., and Kinkley S. PHF13 a novel player in RNA Polymerase II regulation. 2017. Transcription.
Stunnenberg HG., International Epigenome Consortium, Hirst M. The international epigenome consortium: A blueprint for scientific collaboration and discovery. 2016. Cell.
Durek P., Nordström K., Gasparoni G., Kressler C., de Almeida M., Salhab A., Bassler K., Ulas T., Xiong J., Glažar P., Klironomos F., Sinha A., Kinkley S., Yang X., Arrigoni L., Amirabad A.D., Ardakani F.B., Feuerbach L., Gorka O., Ebert P., Müller F., Li N., Frischbutter S., Schlickeiser S., Cendon C., Fröhler S., Felder B., Gasparoni N., Imbusch C.D., Hutter B., Zipprich G., Tauchmann Y., Reinke S., Wassilew G., Hoffmann U., Richter A.S., Sieverling L.; DEEP Consortium, Chang H.D., Syrbe U., Kalus U., Eils J., Brors B., Manke T., Ruland J., Lengauer T., Rajewsky N., Chen W., Dong J., Sawitzki B., Chung H.R., Rosenstiel P., Schulz M.H., Schultze J.L., Radbruch A., Walter J., Hamann A., Polansky J.K. Epigenetic reprogramming in memory formation of human CD4+ T cells. In Press. 2016. Immunity.
Kinkley S., Helmuth J., Polansky J., Dunkel I., Gasparoni G., Fröhler S., Chen W., Walter J., Hamann A., and Chung H-R. reChIP-seq reveals widespread bivalency of H3K4me3 and H3K27me3 in CD4+ memory T-Cells. 2016. Nature Communications.
Chung H., Xu C., Fuchs A., Mund A., Lange M., Schubert T., Steage H., Bian C., Dunkel I., Eberharter A., Regnard C., Klinker H., Cozzuto L. Winterpacht A., Di Croce L., Min J., Will H., and Kinkley S. PHF13 is a Molecular Reader and Transcriptional Co-Regulator of H3K4me2/3. 2016. eLife.
Perner J., Lasserre J., Kinkley S., Vingron M., Chung HR. Inference of interactions between chromatin modifiers and histone modifications: from ChIP-Seq data to chromatin-signaling. 2014. Nucleic Acid Research.



