2. Transposable Elements and RNA Binding Protein networks in post-transcriptional gene regulation
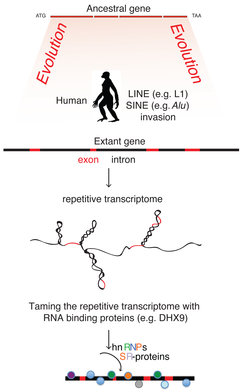
The struggle to keep transposons under control has spawned a large number of cellular strategies that reach beyond their immediate roles in the suppression of Transposable Elements (TEs). The fight against double-stranded RNA, a sign of viral invasion, for example, has likely led to the evolution RNAi, miRNA and piRNA pathways. In higher eukaryotes, selfish elements like LTR, LINE retrotransposons and integrated proviruses are vehemently suppressed by a group of transcription factors that recruit the master regulator KAP1 to repress transcription from these loci (Rowe et al. 2010). These proteins inevitably create a nuclear environment that affects how the rest of the genome is regulated. This becomes especially important for protein-RNA and protein-DNA interactions, which usually involve the recognition of rather short sequence motifs (3 to 6-mers) that are unlikely to be unique to TEs. On the post-transcriptional side of events, our work with DHX9 has shown that DHX9 targets long dsRNA and unwinds them to allow proper RNA processing, which involves splice site choice and cleavage of pre-mRNA to terminate RNAPII transcription.
Ultimately, we would like to find novel RNA-binding proteins that post-transcriptionally target transposable elements, and study their role in the post-transcriptional regulation of normal cellular gene expression in addition to their roles in suppressing the proliferation of TEs, similar to DHX9, hnRNPC, MOV10. We developed a patent-pending UV-crosslinking and sequencing (CLIP-seq) method (which we called uvCLAP (Maticzka et al. 2017) and FLASH ((Ilik, Akhtar, and Aktas-Ilik 2018) and manuscript in preparation)) that will allow us to profile hundreds of RNA-binding proteins within several months. Such speed and throughput is currently impossible with commonly used CLIP-seq methods due to cumbersome, gel-purification based-protocols, however is within reach using our method (Aktaş et al. 2017; Ilik, Akhtar, and Aktas-Ilik 2018).
These data will also be instrumental to uncover the ribonucleoprotein (RNP) composition of low-complexity RNA aggregates in neurodegenerative diseases, and will provide clues to prevent the formation or to promote the dissolution of these aggregates by pharmacologically targeting key factors in this process.
References:
KAP1 controls endogenous retroviruses in embryonic stem cells, Rowe H.M.; Jakobsson J.; Mesnard D.; Rougemont J.; Reynard S.; Aktas T.; Maillard P.V.; Layard-Liesching H.; Verp S.; Marquis J.; Spitz F.; Constam D.B.; Trono D.; Nature, 463, 7278, 237-240, 2010
uvCLAP: a fast, non-radioactive method to identify in vivo targets of RNA-binding proteins Maticzka D.*, Ilik I.A.*, Aktas T.*, Backofen R., Akhtar A.; Nature Communications, 9(1), p.1142, 2018
*equal contribution
DHX9 suppresses RNA processing defects originating from the Alu invasion of human genome Aktas T.*; Ilik I.A.*; Maticzka D.; Bhardwaj V.; Rodrigues C.P.; Manke T.; Backofen R.; Akhtar A.; Nature,544 (7648), 115-119, 2017
*equal contribution
CLONING OF SINGLE-STRANDED NUCLEIC ACID
IA Ilik, A Akhtar, T Aktas-Ilik - US Patent App. 15/744,938, 2018
