Research
The Use of PSCs for Understanding NSC type heterogeneity of the developing human cerebral cortex
Our lab has made a major effort to address heterogeneity in cultured human NSCs. We are particularly interested in understanding the biology of NSCs that function during development of the human neocortex. We use human PSCs in combination with state of the art stem cell culture and differentiation techniques, transgenic methods, transcriptional profiling, quantitative live cell-imaging, extensive epigenetic analyses, and gene loss of function studies – all towards dissecting the establishment, patterning, maintenance and progression of early cortical NSCs derived from PSCs at the cellular, molecular and cytoarchitectural level.
Isolation of Notch-dependent consecutive building blocks for human cortical development
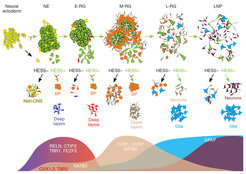
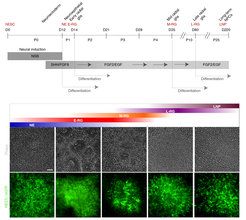
In our first endeavor, we derived a novel type of early neuroepithelial stem cell from PSCs (Elkabetz et al., G&D 2009), which became a platform for our subsequent discoveries. To get a systematic overview of these early NSCs and their progeny in the lab, we used a PSC reporter line for Notch activation, which reports on the existence of NSCs. Using this reporter, we were first able to re-generate and isolate our formerly derived early neuroepithelial stem cells based on their high Notch activation (GFP), and demonstrated their Notch-dependent proliferative capacity and broad developmental potential. Using these cells as a starting population, we consecutively derived and isolated four additional novel types of NSCs through lineage relationship via Notch activation.
Remarkably, we found that NSC types consecutively derived in progressive culture based on their Notch can together recapitulate the ordered generation of cortical layer formation. Thus, similar to development in vivo, each isolated consecutive building block can give rise to its corresponding cortical neuronal layer as well as subsequent progenitor stage. In addition, production of neuronal progenitors preceded emergence of glial progenitors, and these latter progenitors further progress to adult-like NSCs. Importantly, all these processes depended on Notch activation. These findings demonstrate our ability to deconstruct and reconstruct human cortical development in the petri dish with Notch-dependent building blocks.
Our approach showed ability to dissect stem cells from non-stem cells during long-term culture - which allowed for the first time to redefine NSC types and relate them to distinct fate potentials. The finding that NSC stages with distinct potencies can appear in culture despite the consistent culture conditions is an evidence for an intrinsic developmental clock that perpetuates in cultured NSCs, which can be deciphered by studying the molecular changes in these new populations.
For reading:
Elkabetz et al., Genes & Development, 2008.
Edri, Yaffe et al., Nature Communications 2015.
Isolation of consecutive building blocks for cortical development from human pluripotent stem cells based on their Notch activation state. From Edri, Yaffe et al., Nature Communications 2015.
Dissecting cortical differentiation regulatory networks through extensive epigenetic analysis
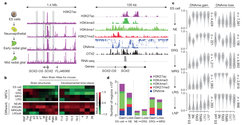
We further took advantage of our cortical progenitor building blocks to uncover the molecular networks that drive cortical differentiation in vitro. Collaborating with the group of Prof. Alexander Meissner at the Broad Institute, we extensively analyzed these consecutively isolated cortical NSC types at the transcriptional and epigenetic level. Using advanced biochemical methods and sophisticated computational frameworks, we detected genomic regions that undergo differential DNA methylation and/or chromatin modification during the cortical differentiation process, and which therefore are suspected to play a role in either establishment of early NSCs of the developing cortex or their transition through the consecutive progenitor stages. Further exploiting these computational frameworks, we uncovered unique sets of transcription factors and epigenetic modifiers that are likely to bind these regions and change the epigenetic landscape in order to either promote, maintain or exit each stage of the differentiation. This resulted in uncovering a core set of constitutively active transcriptional regulators that interact with stage specific transcription factors in a stage dependent manner - to drive transition through NSC stages. We also validated these factors functionally using a large-scale shRNA screen for Notch activation - together leading to the uncovering network that govern establishment, commitment and transition through specific NSC types involved in corticogenesis.
These findings demonstrate the utility of our system to provide new genomic information that can be harnessed for generating more homogeneous NSC populations in the near future. As such, these findings should have far reaching implications for building complex brain networks in the Petri dish for basic and regenerative research and future applications.
For reading:
Ziller, Edri et al., Nature 2015.
Edri, Yaffe et al., Nature Communications 2015.
Quantitative live imaging of cortical NSCs as novel readout for cytoarchitectural dynamics during cortex development
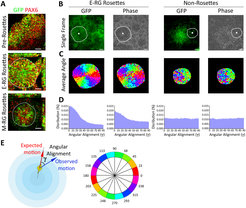
We also aimed at exploring how the establishment and maintenance of cortical NSCs may be reflected – or affected – by their cytoarchitectural dynamics. More specifically, we asked to what extent critical capabilities of NSCs such as self-renewal and differentiation are reflected by kinetic and morphological properties that can be live imaged and quantified.
To objectively measure these dynamics, we developed quantitative live imaging approaches and algorithmic frameworks to characterize cellular and nuclear motion dynamics within these cortical neuroepithelial cell structures – which reflect cortical cytoarchitecture. We particularly investigated how migration patterns of nuclei from apical site to basal site (a process termed inter kinetic nuclear migration, or INM) are associated with NSC state and numbers during cortical development in vitro. We defined objective measures for these INM dynamics such as the nuclei speed, nuclei angle orientation and the ratio of apical vs. basal nuclei direction. We found high performance with respect to these measures, which correlates well with early stages of development in vitro, while observing significant decline upon progression of NSCs towards later stages (as identified in Edri et al.).
This suggests, for the first time, a functional link between neuroepithelial cell dynamics and NSC competence during the building of the cortex. These findings propose that our kinetic quantitative readouts can potentially serve as a platform for functional molecular studies and drug screening, and indeed these readouts are already implicated in our lab for gaining novel kinetic nuclei dynamics insights into the pathology of NSCs during the onset of cortical diseases such as Microcephaly. In addition, and along with our initial long-term goal to generate perpetuating neuroepithelial stem cells, we use these measures constantly as additional hallmarks for assessing the developmental stage of NSCs in culture.
For reading:
Ziv, Zaritsky, Yaffe et al., PLoS Computational Biology 2015.
Generating homogeneous early cortical rosettes and cerebral organoids: Implications in modelling cortical development and microcephaly
We continuously take advantage of the molecular aspects of heterogeneity which we have observed during our studies with early cortical neuroepithelial stem cells for developing improved differentiation protocols for deriving more homogeneous cortical progenitor populations. Our recent data revealed that early neuroepithelial cells are heterogeneous with respect to their CNS regional identity. As it turns out, neural stem and progenitor cells derived by many of the current protocols hold mixed regional identity not necessarily confined to the cortical domain, but are with rather general forebrain fates. Our transcriptional and epigenetic datasets highlighted the involvement of specific pathways that determine regional heterogeneity of the cortex. Based on these, we devised an optimized protocol for combinatorial modulation of WNT, TGF and BMP pathways to streamline and accelerate derivation of homogeneous cortical stem cells with dorsal cortical identify from PSCs.
We confirmed this method for different PSC lines and for a variety of neural differentiation schemes including rosettes and cerebral organoids. Importantly, applying our protocol on cerebral organoids cleared many of the undesired outcomes of the original organoid formation method, overall enhancing the system for obtaining highly reliable data for modelling normal and pathological corticogenesis.
To exemplify the utility of the new protocol in disease modelling, we generated a microcephaly PSC cell line by introducing a genetic defect that causes human autosomal recessive microcephaly by CRISPR technology. Among the many phenotypes we could record, we importantly were able to measure cortex-specific cell death in our derived organoids that was not apparent by other derivation protocols. This enabled us to delineate a novel early pathology of microcephaly in cortical NSCs. These results further emphasize the critical role of reduced heterogeneity in disease modelling.
Taken together, we envisage that our streamlined protocol marks an important step towards generating more clinically relevant and purified cortical NSCs and their neurons - for studying basic and regenerative aspects of their biology. In addition, increased homogeneity in starting populations is expected to ease molecular understanding of NSCs involved in the disease.



