Edda Schulz receives prestigious funding from the European Research Council
ERC Starting Grant to help explain stable gene expression
The biophysicist Edda Schulz from the Max Planck Institute for Molecular Genetics (MPIMG) in Berlin was awarded an ERC Starting Grant of 1.5 million euros. The funding programs by the European Research Council are among the most important in Europe. Schulz is investigating how cells integrate information from different sources and use this information to make developmental decisions.

It is a massive undertaking: During individual development, an entire organism with millions of different cells has to rise from a single fertilised egg cell. Its blueprint – the genome – is however the same in each cell. So what determines which cell becomes part of the brain, the heart or the belly button – and makes sure that it stays put permanently?
“On the one hand, every cell must react dynamically, to hormones, environmental cues or altered gene activity in the cell itself; but on the other hand, it must retain its identity," says Edda Schulz, head of the Lise Meitner Research Group “Systems Epigenetics” at the MPIMG. She has recently received project funding of 1.5 million euros from the European Research Council (ERC). Over the next five years, Schulz hopes to elucidate why some genes remain active over long periods of time, while others remain permanently turned off.
Here to stay – for a lifetime
Schulz uses the X chromosome as a model for the fundamental question of how gene activity is stabilized. The X chromosome is one of two types of sex chromosomes. Cells of female mammals usually have two copies of the X chromosome, male cells have one X and an additional Y chromosome.
“Even though females have two of them, every mammal needs only one active X chromosome per cell,” explains Schulz. “One of the two chromosomes has to be switched off, permanently for life and irreversibly in every somatic female cell. The chromosome that is inactivated is chosen by chance.” The other chromosome must also remain permanently active, because it contains a large amount of essential information that is needed for reproduction, brain development and cognitive processes like learning and perception. Five percent of known human genes are found on the X chromosome.
The “surplus” chromosome gets inactivated early during development, when the embryo consists of only a handful of cells. At some point, the Xist gene (pronounced “exist”) awakens on only one of the two X chromosomes. Then, numerous RNA copies of Xist are produced that envelop the chromosome. An armada of epigenetic factors follows that flag the DNA as inactive and cause the chromosome to shrink into a tiny lump.
Cells do not throw dice and cannot count
But how does the cell choose between the two X chromosomes to inactivate one of them? How does it know that it should remain active in male cells?
“Cells can neither throw dice nor count,” says the scientist. “Instead, nature makes use of biochemical and molecular circuits.” These consist of genes and RNA as well as protein molecules that can dock to structural features in the cell.
“X-chromosome inactivation is a very complex process that depends on the timing of development and the biological sex of the cell,” says Schulz. For it to succeed, the cell has to integrate information from its immediate vicinity, from the cell's own genome and on the number of X chromosomes.
Integrating cellular signals – globally and locally
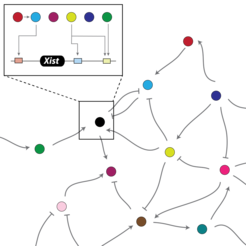
In principle, one X chromosome must remain active in every cell, regardless of whether it is male or female. The Xist gene, which is responsible for X inactivation, must therefore initially be inactivated itself. However, if there are two X chromosomes in the cell, Xist will be activated and will silence one of them. This is achieved by molecular switches in the immediate vicinity of the Xist gene, which only exert their effect locally on the chromosome on which they themselves are located. These switches are again actuated by other X-chromosmal factors, but these also affect genes that are not on the same but on another chromosome.
Taken together, it means that with only a single X chromosome present in a cell, its inactivation will be prevented and Xist should not be transcribed. If there are more X chromosomes, however, Xist kicks in and the X inactivation cascade starts. A number of other local molecular switches further complicate this interplay of factors.
Understanding the network of molecular circuits
“We want to figure out the role of all the factors and how they work together – and to what extent each of them is involved,” says Schulz. She and her team want to reconstruct the network of genes, regulatory RNAs and other molecules as a mathematical model in the computer. But first, they will have to test which DNA segments regulate which gene.
The experimental system is the culture of embryonic stem cells of mice. Using CRISPR-Cas9, Schulz and her team will specifically manipulate DNA segments. By sequencing the transcribed RNA of each and every cell and thus determining the activity of each DNA segment, it will be possible to track the effects of each genetic alteration.
This approach enables the quantitative observation of cellular processes. As with determining the number of X chromosomes, some processes are apparently only triggered once enough factors have accumulated, says Schulz: “Sometimes it needs many people pushing to get the ball rolling.”
Schulz is therefore not only interested in reconstructing mechanisms, i.e. finding out the sequence of events in the cell, but quantifying them, too. She believes that many regulatory functions depend on the rates of formation and degradation processes and corresponding threshold values.
An exemplary section from the overall picture
Compared to the overwhelmingly complex network of processes that comprises a cell or even in the entire organism, the action of the Xist gene is certainly only a very small part. However, because X-chromosome inactivation is a robust mechanism that is not dependent on most cellular processes, it is comparatively easy to study. At the same time, X-chromosome inactivation makes use of the general molecular toolbox of the cell and can therefore be viewed as an example of similar workings in the genome.
“Our ultimate goal is to understand gene regulation over several levels of complexity,” says the scientist. Together with the additional funding from the ERC, she has the ideal conditions for her vision at the MPIMG.

About Edda Schulz
After completing her doctorate as a biophysicist at the German Rheumatism Research Centre Berlin (DRFZ) and at the Humboldt Universität zu Berlin, Edda Schulz won one of the prestigious long-term fellowships of the Human Frontiers Science Program in 2010 and joined Edith Heard's group at the Institut Curie in Paris as a postdoctoral fellow. In 2014, she established a Max Planck Research Group at the MPIMG, where she is currently heading a team of ten researchers. Most recently, Schulz was able to secure a Lise Meitner grant from the Max Planck Society.
About the ERC
The European Research Council, set up by the European Union in 2007, is the premiere European funding organisation for excellent frontier research. Every year, it selects and funds the very best, creative researchers of any nationality and age, to run projects based in Europe. The ERC offers four core grant schemes: Starting, Consolidator, Advanced and Synergy Grants. The ERC strives to attract top researchers from anywhere in the world to come to Europe.















