Yaspo lab projects
Metastatic Melanoma (Treat20Plus)
(Funded by the German Ministry of Education and Research)

Treat20, was a pilot translational project designed to predict the drug response for 20 metastatic melanomas based on their molecular features and on the prediction of drug response (Christoph Wierling’s group). In Treat20, we analysed several melanoma types (cutaneous, uveal, iridal, mucosal and acrolentiginous), and found novel molecular events in non-UV induced, non-BRAF mutated melanomas, providing potential therapeutical targets. Treat20Plus, is a follow up project designed as a clinical trial for 35 metastatic melanomas refractory to treatment. The project is coordinated by ML Yaspo, in partnership with the CCCC, Charite (Prof. Ulrich Keilhoz) and Alacris Theranostics carrying out the tumor modeling for treatment recommendation. Our group will carry out the deep NGS analysis and will establish melanoma-derived microtissues, providing a platform for drug testing and model validation, but also a tool for investigating the interactions between melanoma cells and their stromal environment.
Individualized Paediatric Cure (iPC): Cloud-based virtual-patient modules for precision paediatric oncology

The objective of the iPCs network is to improve care for children with cancer by developing cloud-based technologies and infrastructures to help identify effective individual therapies. The project team include leading paediatric cancer researchers and clinicians who will focus on the integration of high-quality data sources and their analyses using both knowledge-based and artificial-intelligence models with the goal of boosting the power of each dataset and improving the therapeutic decision making of paediatric cancers. To achieve this goal, iPC will lay the technical basis for the development of the virtual patient framework – a comprehensive and personalized patient model that can facilitate the evaluation of many different treatment options to maximize the balance between treatment efficacy and toxicity, thus improving both patient survival and their quality of life. Furthermore, these in silico generated virtual patient models will be evaluated for their predicitve ability and faithful patient representation using preclinical models, biochemical assays, and through prospective testing within ongoing clinical trials.
The Yaspo group is supporting the iPC project by conducting the gene perturbation screens and gene-drug interactions for mechanistic model development. Our efforts are focused on a subset of childhood tumor types: paediatric leukemias, medulloblastoma, Ewing sarcoma, hepatoblastoma and neuroblastoma. For each paediatric tumor type, we perform CRIPSR-Cas9 perturbation screens to evaluate thousands of identified genes associated with tumors. Generated gene expression phenotypes are then assessed by single-cell RNA sequencing readout. Such combination of both methods, determines the gene expression signatures for individual knockouts in massive parallel fashion, studying thousands of pooled cells in a single experiment. Therefore, we are able to identify genes involved in biological mechanisms (i.e. cell proliferation and drug resistance) and provide insight into complex signaling pathways in heterogeneous cell populations. The data generated in all experiments will be further used for analysis of network structure and parameter optimization for mechanistc models.
3TR: Accelerating Precision Medicine for Immune-Mediated Diseases

The lab participates in the 3TR project that aims to provide fundamental new insights into the mechanisms of response and non-response to treatment. It is a large research project funded by the Innovative Medicines Initiative 2 (IMI2) that will provide important new insights and information about why a large number of patients suffering from the following seven diseases do not respond to treatment: asthma, COPD, Crohn’s disease, ulcerative colitis, multiple sclerosis, systemic lupus erythematosus, and rheumatoid arthritis.
Multimodal Clinical Mass Spectrometry to Target Treatment Resistance (MSTARS)
Our group is part of the Berlin research core MSTARS that also includes groups from Charité and the Max Delbrück Center, as well as scientists from the Berlin Institute of Health and Humboldt-Universität zu Berlin, gathering all Berlin-based expertise in the fields of mass spectrometry, patient care and data analysis, and placing it under one roof.
The aim of the MSTARS consortium is to further develop current technology for clinical application and for the detection and classification of treatment resistance. In order for this technology to improve patient care, we also need to be able to analyze large numbers of samples within a short time. To achieve this, we will build capacity and develop standardized procedures for all processes, from sampling through to data management.
The idea behind the project is to combine the skills and experience of the various institutions and disciplines with Charité’s extensive clinical expertise to enable development of mass spectrometry-based technologies and apply them in clinical practice.
MSTARS (Multimodal Clinical Mass Spectrometry to Target Treatment Resistance) is being funded under a special BMBF funding program, which focuses on establishing research cores dedicated to mass spectrometry in systems medicine (‘Forschungskerne für Massenspektrometrie in der Systemmedizin’).
TREGeneration: Technological developments for the analysis of deep immune status
TREGeneration – EU Horizon 2020 Research and Innovation Programme

The analysis of immune cell repertoires using NGS techniques, is carried out by Dr. Hans-Jörg Warnatz.
We have developed a patent-pending technique for high-throughput paired amplification of antibody-encoding heavy and light immunoglobulin (Ig) chains from B cell populations ("PairedImmune" sequencing, International patent application PCT/EP2013/052328). The information on natively paired antibody-coding Ig gene pairs from populations of single B cells can be used for the generation of high-affinity antibodies useful as dignostics and passive vaccines. We have successfully applied this technique to peripheral B cells from a healthy donor, and we are now analyzing B cell populations from a healthy donor before and after a Tetanus vaccine boost.
In the new European "TREGeneration" project, has its goal to develop new strategies within clinical trials for the treatment of Graft versus Host Disease, a serious complication following bone marrow transplantation, our task is to characterize the T cell immune status of the patients and donors at key time points of the clinical trials.
Completed projects
International Cancer Genome Consortium
Pediatric brain tumors (ICGC Pedbrain)
We participate in the PedBrain Tumor Project,which is part of the International Cancer Genome Consortium (ICGC) and focuses on pediatric brain tumors.
Transcriptome analysis of large pediatric brain tumor cohorts
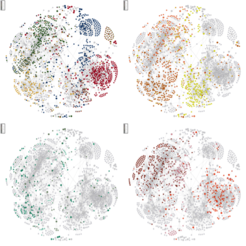
As part of the International Cancer Genome Consortium (ICGC), in ICGC Pedbrain,we generated RNAseq data and analysed the transcriptomes of 164 medulloblastomas (MB), 90 pilocytic astrocytomas (PA) and 50 glioblastomas (GBM).
MB is the most common malignant brain tumour in children arising in the cerebellum or medulla/brain stem and shows biological and clinical heterogeneity. Previous studies idnetified four main groups of MBs with distinct clinical, biological and genetic profiles. WNT tumours, associated to a favourable prognosis, characterized by activated wingless pathway signalling, tumours showing hedgehog pathway activation with intermediate prognosis, and group 3 and 4 tumours, that are less well characterized and clinically challenging.
Our group identified the first medulloblastoma-associated gene fusion, involving the SHH gene (Jones et al. 2012). We identified gene expression signatures specifying nine different MB sub-groups either reflecting their cells of origin or re-programming of the tumor cells and identifying novel pathways . In group 3, we found one cluster enriched for rhodopsin and markers associated to visual perception, one myc-driven cluster, and one that is expressing a set of early develomental genes. We are particulary interested in the sub-group specific transcriptional networks.
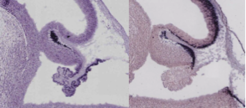
Pilocytic astrocytoma (PA) accounting for ~20% of all pediatric brain tumors, is typically associated with mitogen-activated protein kinase (MAPK) pathway alterations. By integrating genome and transcriptome information surveying a cohort of 96 tumors, we demonstrated that PA is indeed a single pathway disease, featuring novel gene genes fusions involving the RAS pathway, such as BRAF or the kinase domain of the NTRK2 oncogene (Jones et al. 2013).
ICGC Early Onset Prostate Cancer
As part of the International Cancer Genome Consortium (ICGC), we are contributing to the EOPC consortium, in particular with RNAseq analyses of the tumor cohort.
IMI Oncotrack Colorectal carcinoma

Colorectal carcinoma (CRC) is the third most common cancer worldwide and the second most common cancer in Europe, representing a major healthcare burden. KRAS mutations are used as predictive marker of therapeutical response to antibodies targeting EGFR, however, the resistance to therapeutics remains poorly understood. The IMI Oncotrack international project (www.oncotrack.eu) aims at analysing a prospective CRC cohort with „multi-omics“ technologies for identifying novel biomarkers of disease and drug response.
The Yaspo group is coordinating the genomic and transcriptomic analysis of the CRC cohort and matching patient-derived models. Together with the spin-off Alacris Theranostics, we have analysed, whole genome, exome, microRNA and RNAseq data of more than 100 tumors, 60 xenografts and 40 spheroid cell models. Based on these data, our main research directions focus on:
- Identifying specific signaling pathways in CRC sub-groups
- Dissecting tumor heterogeneity and clonal evolution in tumor models
- Correlating drug sensitivity of the models with molecular biomarkers
Besides hallmarks mutations in APC, Wnt signaling and RAF/RAS pathway, we identified novel somatic changes in chromatin remodelling complex and epigenetic factors, which we are currently investigating,. We observed significant intra-tumoral heterogeneity, reflected in pre-clinical models of the tumors often displaying a different molecular setup than their donors. Further, expression of stem cell markers was significantly different in the models, either due to epigenetic reprogramming and/or to clonal evolution. We are investigating this in cooperation with Mats Nielson, (Uppsala, Sweden) who is using a combination of multi fluorescent labelling and in situ hybridisation.
UFOPLAN – Pediatric acute lymphocytic leukemia ALL
(Funded by the German Federal Office for Radiation Protection)

Integrated genomic and functional analyses of TCF3-HLF-positive ALL
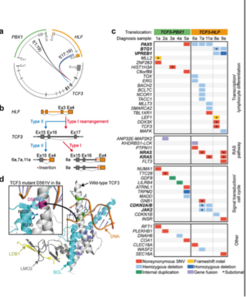
Pediatric acute lymphocytic leukemia (ALL) is characterized by chromosomal translocations that cause gene fusions involving master regulators of hematopoietic development. In a collaborative project, we aimed at comparing the molecular pathology between two different leukemia types both disrupting one allele of TCF3, which drives the B-cell differentiation program upstream of PAX5.:the t(1;19) fusing TCF3 to the DNA-binding domain of PBX1 associated with a good prognosis.and the dismal t(17;19)(q22;p13), resulting in the fusion TCF3-HLF.
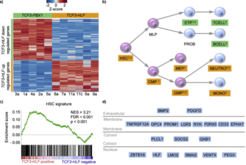
The two leukemia subtypes shared a gene expression signature of B-lymphoid cells, but also showed striking differences.We found that expression of TCF3-HLF leads to transcriptional reprogramming in B lymphoid progenitors in the context of PAX5 haploinsufficiency, towards an immature, hybrid hematopoietic state. TCF3 breakpoints occurred in hotspots indicative of terminal deoxynucleotide transferase (TdT) activity characteristic of an early B cell stage. Using In silico analysis, we predicted 39 potential HLF targets in TCF3-HLF samples, including SNAI2,, GPC4 and BMP3 involved in stem cell proliferation A profound cellular reprogramming occurred in TCF3-HLF towards a drug-resistant state reflected by stem cell, mesenchyme-derived and myeloid signatures. Drug response profiling in patient-derived xenografts, which had maintained the tumor’s genomic and transcriptome landscapes, identified resistance to most of the 98 tested drugs, but extreme sensitivity towards the BCL2-specific inhibitor ABT-199, indicating new therapeutic options for this fatal ALL subtype (Fisher et al. Nature genetics, 2015).

BLUEPRINT - A BLUEPRINT of Haematopoietic Epigenomes
We are also participants of the Blueprint epigenome project. BLUEPRINT is a large-scale research project receiving close to 30 million euro funding from the EU. 42 leading European universities, research institutes and industry entrepreneurs participate in what is one of the two first so-called high impact research initiatives to receive funding from the EU.














