Mapping embryonic development
3D atlas of gene expression offers new insight into tissue and organ development.
The lab of Alexander Meissner at the Max Planck Institute for Molecular Genetics (MPIMG), in collaboration with the Broad Institute and the University of Marseille, has developed 3D “virtual embryos” that allow scientists to study the spatial distribution of gene expression during development. Their work, published in Nature Genetics, reveals details of how tissues develop in the early mouse embryo.
The cellular world can appear two-dimensional to someone looking at it through a microscope. In reality, biological processes in tissues and organs take place in three-dimensional space. The molecular mechanisms occurring in a single cell are just as important as its location within a tissue or organ, and which genes are being actively transcribed at a particular time and place. This is especially important during early development, when the precise timing and location of gene activity orchestrates the specification of various cell types resulting in organ formation. "The embryo is a very fast-forming entity," says Adriano Bolondi, a postdoc in the Meissner lab, and one of the first authors. "It is important to analyze embryonic development from a multidimensional perspective, because it relies on a variety of complex inputs and signaling processes that might otherwise be missed." Recent technological advances have made it possible to study the transcription of genes in their native environment, including within developing tissues. Alexander Meissner and colleagues have taken this approach to the third dimension.
From 2D snapshots to virtual embryos
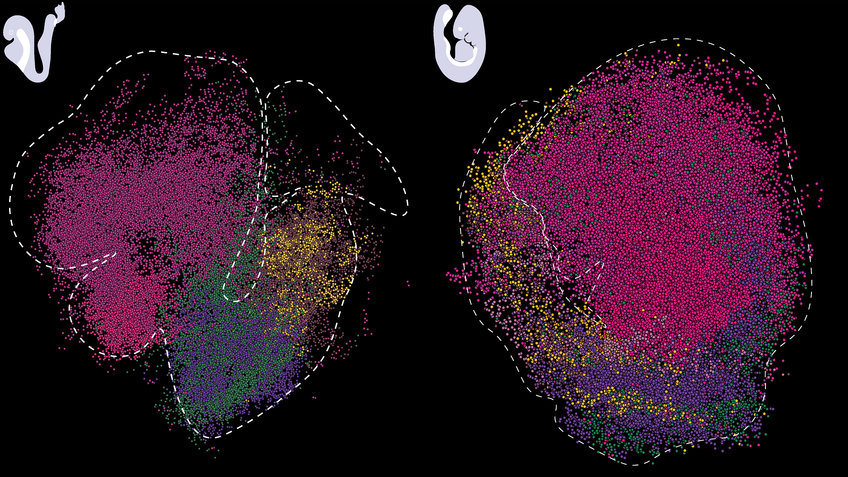
The team used Slide-seq, a method developed by Fei Chen and Evan Macosko’s laboratories at the Broad Institute. The idea behind the technology is simple. First, tissue samples from mouse embryos are collected, each containing a “snapshot” of the gene activity in that particular area. Using microscopic beads labeled with DNA barcodes, the products of gene transcription, RNA molecules, are captured and later sequenced. These data then allow to map the identity and quantity of active genes at each location. "One of the main technological advances of our work is a computational method named sc3D, which we developed together with Léo Guignard, a computational biologist at the University of Marseille. It allows us to align the transcriptomic slides, which are two-dimensional, into a three-dimensional object that we call a “virtual embryo”," says Adriano Bolondi. Along with this approach, the team developed a visualizer that allowed them to interactively explore the expression patterns of about 27,000 genes in the developing embryo.
New insight into nervous system development
The scientists applied this toolkit to study nervous system development in detail. They focused on the posterior neural tube, a structure that later gives rise to the spinal cord and is crucial to generate neuronal diversity. They reconstructed distinct gene expression patterns in this tissue and mapped previously unannotated genes that are restricted to specific parts of the tube. The discovery of these genes raises immediate questions about their molecular roles, which the scientists now plan to further pursue. Finally, they have used their method to characterize the transcriptional landscape of a classic genetic mutant that forms additional neural tubes during embryogenesis, a lethal defect. "Overall, our method increases the granularity with which we can study mammalian development, including when things don’t go as expected," says Adriano Bolondi. The team now hopes that their tool will serve as a valuable resource to ultimately construct a complete map of gene regulation during development and to rationalize perturbations and developmental defects during embryogenesis.