Scientific projects
Understanding the principles of productive TF:DNA interactions
[Verena Thormann, Laura Glaser]
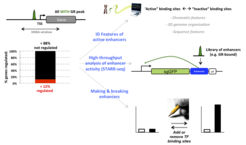
TFs can bind to tens of thousands of genomic binding sites, yet they seem to regulate a much smaller number of genes. We combine genome-wide and focused studies with individual enhancers to unravel molecular mechanisms that facilitate “productive” TF interactions (Fig. 1). For example, we developed a massively parallelized reporter assay to quantify the intrinsic activity of tens of thousands of enhancers. Furthermore, using computational approaches, we identified candidate features (e.g. sequence motifs) associated with active enhancers. To test the relevance of identified features, we will study the effect of their deletion on gene regulation (“reverse engineering”). In parallel, we generate synthetic genomic enhancers (“forward engineering”) to determine if we really understand the operating principles of productive TF binding.
Specificity of related TFs
[Marina Borschiwer, Melissa Bothe]
Paralogous TFs often share similar DNA sequence preferences, yet regulate distinct genes and execute unique functions in vivo. For example, GR and the androgen receptor (AR) recognize essentially the same sequences, yet whereas AR stimulates muscle gain, GR promotes muscle wasting. To address this paradox, we mapped genome-wide binding and changes in gene expression and chromatin landscape in response to activation of either receptor. Furthermore, we mapped the intrinsic enhancer activity of the majority of regions bound by both receptors. Consistent with previous studies, we find that receptor-specific binding can explain some of the differences in gene regulation. Interestingly, we also identified cases where the specificity appears to arise downstream of binding, given that the genomic loci responsible for the regulation of a gene are occupied by both receptors, yet only one is capable of activating the gene. Ongoing efforts aim at understanding what directs this specificity downstream of binding.

DNA as an allosteric modulator of GR structure and activity
[Stefanie Schöne, Marcel Jurk]
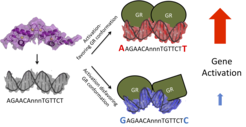
Genes are not simply turned on or off, but their expression is fine-tuned. We study the role of one signal that modulates activity: the exact sequence of the DNA to which GR binds. Computational analysis showed that the nucleotides directly flanking the core-binding site differ depending on the strength of GR-dependent gene activation. Follow-up experiments showed that these flanking nucleotides change the three-dimensional structure of the DNA and of the associated GR protein and that the sequence-induced changes in GR activity cannot be explained by differences in GR occupancy. Rather, mutations that mitigate DNA-induced changes in structure also blunt changes in activity, arguing for a role of DNA-induced structural changes in modulating GR activity downstream of binding.
The role of the genetic and “epigenetic” landscape in guiding GR to specific genomic loci
[Stephan Starick, Jonas Telorac]
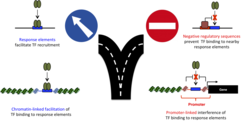
Binding-site motifs of TFs are ubiquitously found in the genome whereas binding only occurs at a subset of putative sites. Whereas most studies focus on the identification of sequences that recruit TFs to specific genomic loci, we identified and characterized negative regulatory sequences that interfere with DNA binding of GR. Contrary to our expectation, we found that these sequences exert their effect by mechanisms other than chromatin accessibility, possibly involving anchoring to sub-nuclear regions that are less permissive to TF binding [Telorac et al., Nucleic Acids Res 2016]. In addition to sequence, chromatin plays a key role in specifying the genomic binding pattern of TFs. One interesting and surprising finding we made was that GR binding is depleted at promoter regions. Sequence composition can explain part, but not all, of this depletion indicating that active mechanisms exist that prevent GR from binding at promoter-proximal regions.
Linking GR binding to the transcriptional regulation of genes
[Verena Thormann]
Using genome-editing and by integrating information regarding chromatin state and the 3D organization of the genome, we identified features that distinguish “productive” (resulting in the regulation of a gene) from “non-productive” GR binding events. For example, the simultaneous presence of a cluster of GR binding sites is required for the activity of certain enhancers. Further, by deleting GR binding sites that are shared between different cell types; we found that cell type-specific genome organization and enhancer-blocking can result in cell type-specific wiring of promoter-enhancer contacts. This rewiring allows an individual GR binding site to direct the expression of distinct transcripts and thereby contributes to the cell type-specific consequences of glucocorticoid signaling.



