Research
Our projects
Regulatory sequence motifs and epigenetic regulation
[Xinyi Yang, Anna Ramisch, Tobias Zehnder, Verena Heinrich, Philipp Benner]
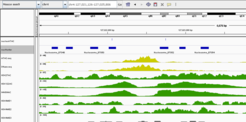
Work is continuing on regulatory sequence motifs and epigenetic regulation. One project provides an integrated analysis of promoter patterns together with ChIP-seq data for transcription factors and histone modifications. We find distinct groups of promoters characterized by certain TF combinations as well as promoters to which CTCF binds and apparently mediates promoter-enhancer looping. At the same time, we are developing machine learning methods to predict enhancers from epigenetic marks. Based on correlation to genes within the same topologically associated domain, these predicted enhancers are subsequently linked to potential target promoters/genes. In the future, hidden Markov models shall serve to integrate information about chromatin into a coherent picture about the regulatory state of a cell type or tissue.
Sequence analysis
[Hossein Moeinzadeh, Stefan Haas, David Heller, Giuseppe Gallone]

Following our earlier work on sequencing and phasing, and mutation identification, the Mundlos and Vingron groups have joined their efforts in genome analysis. One project deals with the mole Talpa occidentalis which is a peculiar model organism for studying skeletal and gonadal development because the female moles develop ovaries as well as a testis-like tissue. We constructed an annotated draft genome assembly covering >17000 known genes. Genomic comparison with human and mouse revealed a triplication of a key enzyme involved in testosterone synthesis and we discovered a small group of genes involved in early development of kidney specifically expressed in the female testis. On the human side, we are currently focusing on the exploitation of long-read sequencing technologies for detection of structural variations in human individuals. In the long run, we aim at combining diverse technologies for the purpose of extensive characterization of individual genomes.
Organoids and single cell transcriptomics
[Ela Gralinska, Lam-Ha Ly, Virginie Stanislas]
In collaboration with Y. Elkabetz from the Meissner department, we are analysing transcriptomes of brain organoids. New methods are being developed to compare publicly available tissue transcriptomes to the ones generated from organoids as well as their individual single-cell transcriptomes. For this purpose, we are currently exploring normalization and visualization methods (mostly correspondence analysis) that allow us to bridge from single cells to organoids to tissues.

