Results of completed projects (selected)
Haplotyping
[Hossein Moeinzadeh, Stefan Haas, Jun Yang]

In collaboration with a Chinese plant biology group we have sequenced a hexaploid plant, the sweet potato Ipomoea batatas, with most of the DNA sequencing done at MPIMG. While it was generally assumed that phasing such a complicated genome would be impossible, it turned out that this particular genome is very heterozygous and even short Illumina sequencing reads contain sufficient polymorphisms to attempt phasing. In the group, we developed the necessary algorithms for phasing and succeeded in assigning almost half of the genome to haplotypes.
3D Chromatin structure
[Robert Schöpflin, Verena Heinrich, Stefan Haas]
We have established a close collaboration with the group of Stefan Mundlos focusing on 3D chromatin structure. Schöpflin, Heinrich and Haas developed tools and provide data analysis support for analyzing 3D chromatin structure data with respect to implications for gene regulation. The emphasis lies on enhancer-promoter interactions, which are affected due to genomic rearrangements with subsequent changes in chromatin interactions.
Sequence determinants of CpG-islands
[Matthew Huska]
We designed a classification method that predicts non-methylated islands (NMI) in different vertebrates. While in human and mouse, the information comes from CpG islands, in other organisms CpG dinucleotides are not sufficient to predict non-methylated islands. Rather, the NMIs in other organisms are defined by more complex patterns, which nevertheless allow for a good prediction of NMIs.
PWM enrichment statistics
[Wolfgang Kopp]
Motif enrichment analysis (MAE) is a frequently occurring task, where one searches for sequence motifs which are enriched, e.g., in a set of jointly regulated promoters. The corresponding statistical problem lies in determining the distribution of the number of hits of a motif in a long sequence. Based on work of the group done earlier [Pape et al., J Comp Biol 2008], we have now further developed this statistics and can compute extremely accurate p-values even under higher-order Markovian background models. This development establishes a sound basis for a hitherto very heuristic method.
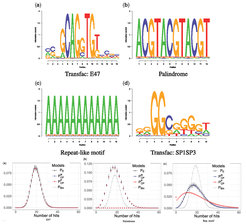
Reconstruction of biological networks
[Mahsa Ghanbari, Julia Lasserre, Ercan Kuruoglu, Alena van Bömmel, Edgar Steiger]
On the theoretical side of the biological networks, several new algorithms were developed. One algorithm is devoted to the question of integrating prior knowledge about gene interactions into the network reconstruction algorithm. Another method aims at delineating changes in network connectivity, when studying time-course expression data. Lately, we have worked on developing a new approach to network reconstruction that is based on “distance correlation” rather than Pearson correlation, which has the advantage that no linearity assumptions need to be made. We developed the DPM (Distance Precision Matrix) Method, which allows for gene network reconstruction even for non-linear relationships among gene. Networks of interactions among transcription factors in enhancer regions are the focus of another new method, which determines in a tissue-specific manner, which pairs of transcription factor binding motifs characterize the enhancers of that particular tissue. This was applied to provide a comprehensive analysis of regulatory interaction in mES cells.

Transcriptome analysis of lung tumors
[Stefan Haas, Ruping Sun]
In a collaborative project led by Roman Thomas, University Cologne, we analyzed RNAseq samples of five lung tumor types with special emphasis on small-cell lung cancer (SCLC) and large-cell neuroendocrine carcinoma (LCNEC). Despite being histologically very similar the clustering of expression data of SCLC and LCNEC not only showed major differences in gene expression between this tumor types but also revealed two subgroups of LCNEC samples. We also found several tumor-type specific splice isoforms, including recurrent splice variants of transcription factors TP73 and E2F7, which are involved in cellcycle
regulation. In an additional project on SCLC patient-derived xenografts derived from circulating tumor cells we predicted candidate fusion transcripts along multiple passages of xenografts. This analysis showed that key fusion transcripts are robustly expressed even after several xenograft passages.
Regulatory networks in stem cells and reprogramming
[Wolfgang Kopp]
Earlier work on epigenetic regulation and epigenetic networks has continued. In collaboration with the group of A. Valencia we analysed a comprehensive epigenetic data-set for mouse ES cells and determined the network of interactions using the methods that had been developed in our group. Another collaboration with the lab of Hans Schöler focused on the role of Esrrb in reprogramming.


